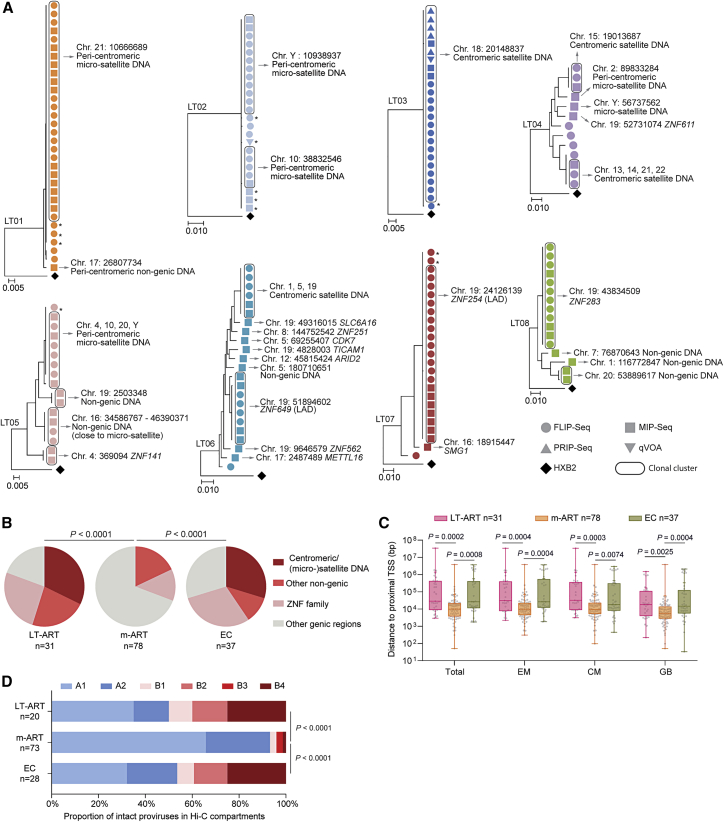
Figure 2.
Chromosomal positioning of intact HIV-1 proviruses in long-term ART-treated individuals
(A) Maximum-likelihood phylogenetic trees for intact HIV-1 proviruses from long-term ART-treated individuals (LT01–LT08). Coordinates of chromosomal integration sites obtained by integration site loop amplification (ISLA) and corresponding gene name (if applicable) are indicated. Symbols indicate sequences generated by FLIP-seq, MIP-seq, PRIP-seq or from quantitative viral outgrowth assays (qVOAs). Proviruses integrated in highly repetitive satellite DNA could not always be definitively mapped to specific chromosomal locations; a detailed list of integration sites is shown in Table S1. ∗Sequences differ by 1 or 2 base pairs from adjacent clonal sequences. LADs, lamina-associated domains.
(B) Proportions of intact proviruses with indicated integration site features in LT-ART individuals and comparison cohorts.
(C) Chromosomal distance between integration sites of intact proviruses to most proximal host transcriptional start sites (TSSs), as determined by RNA-seq in CD4 T cells from reference datasets in total, effector-memory (EM), or central-memory (CM) primary CD4 T cells or from genome browser (GB).16 Box and Whisker plots show median, interquartile ranges and minimum/maximum.
(D) Proportions of genome-intact proviral sequences in structural compartments/subcompartments A and B, as determined by Hi-C seq data.24 Integration sites not covered in the reference dataset were excluded.
(B–D) Data from individuals with moderate ART treatment durations (m-ART) and from EC are shown for comparison. Clonal sequences are counted once. (B–D) p values were calculated by FDR-adjusted two-sided Kruskal-Wallis nonparametric tests or chi-square tests, adjusted for multiple comparison testing where applicable. n reflects the number of integration sites.