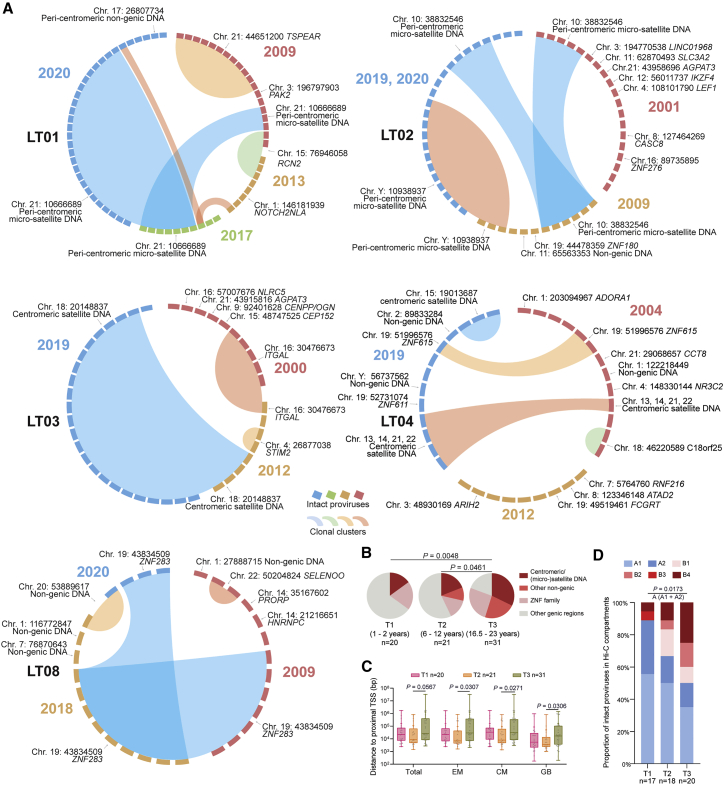
Figure 4.
Longitudinal evolution of intact proviral reservoir landscape during long-term antiretroviral therapy
(A) CIRCOS plots reflecting the chromosomal locations of intact proviruses at indicated time points in five study participants (LT01–LT04, LT08). Each symbol reflects one intact provirus. Clonal sequences, defined by identical integration sites and/or complete sequence identity, are highlighted.
(B) Pie charts reflecting longitudinal evolution of chromosomal integration site landscapes of intact proviruses at indicated time points. Data indicate proportions of intact proviruses in indicated chromosomal locations.
(C) Chromosomal distance between integration sites of intact proviruses to most proximal host TSS, as determined by RNA-seq in CD4 T cells from reference datasets in total, effector-memory (EM), or central-memory (CM) primary CD4 T cells or from genome browser (GB).16 Box and Whisker plots show median, interquartile ranges and minimum/maximum.
(D) Proportions of genome-intact proviral sequences in structural compartments/subcompartments A and B, as determined by Hi-C seq data.24 Integration sites not covered in the reference dataset were excluded. For T3, data from 2017 and 2020 from LT01 were pooled.
(B–D) Clonal sequences are counted once. n reflects the number of integration sites. p values were calculated by chi-square tests in (B) and (D), and by FDR-adjusted two-sided Kruskal-Wallis nonparametric tests in (C), adjusted for multiple comparison testing where applicable.