Figure 2.
Skin MAIT cells accumulate in the wound and constitute a homogeneous type 17 T cell population with a tissue repair program
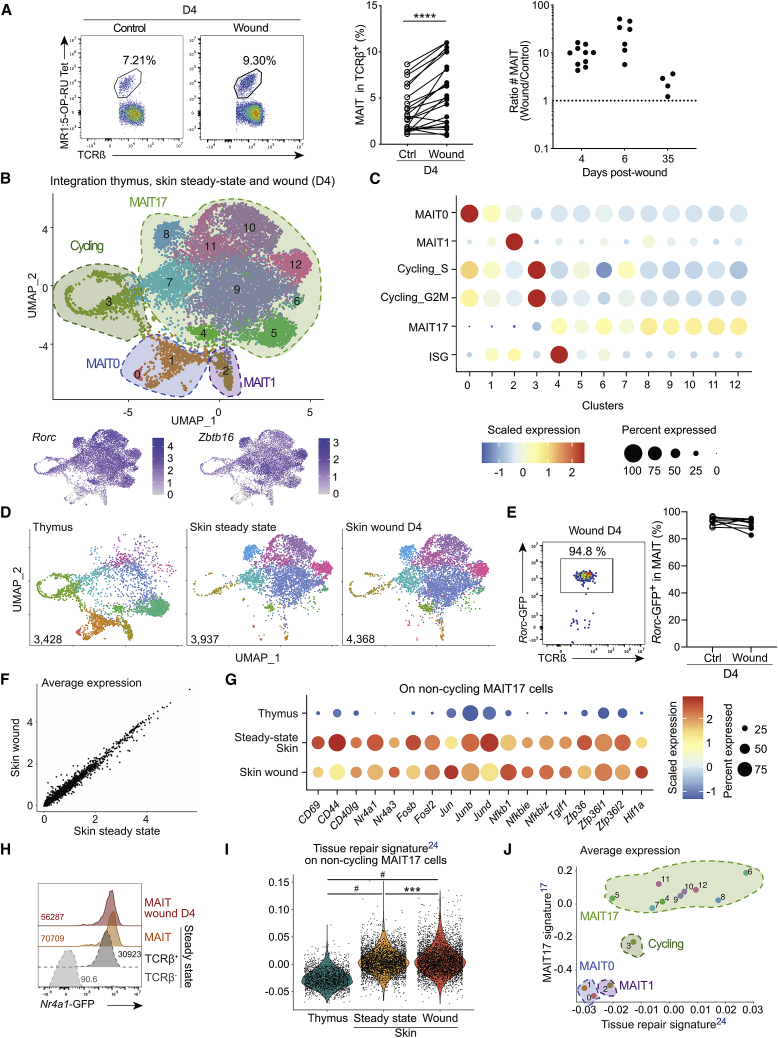
(A) Flow cytometry staining (left), frequency (middle), and number (left, ratio wound over control numbers) of skin MAIT cells from wound and control sites at various time points. Pooled data from seven (n = 22) and four (nD4 = 10, nD6 = 7, and nD35 = 4) independent experiments for frequencies and numbers, respectively. Wilcoxon test.
(B) MAIT cells sorted from thymus, wound (D4), and steady-state skin were analyzed by scRNA-seq and integrated. UMAP (Uniform Manifold Approximation and Projection, top) and features plot for Zbtb16 and Rorc expression (bottom) are displayed.
(C) Cluster were defined by signature enrichment (Table S1; STAR Methods).
(D) UMAP from (B) split according to dataset origin.
(E) Rorc-GFP reporter expression by MAIT cells from wound and control sites. Pooled data from three independent experiments (n = 8). Please also see Figure S2B.
(F) Average gene expression from MAIT cells in wound site and steady-state skin.
(G) Differentially expressed genes in non-cycling MAIT17 cells from skin (wound and steady state) as compared to thymus. The average expression was calculated on scaled data after subsetting MAIT17 clusters from (B).
(H) Nr4a1-GFP reporter expression by skin MAIT cells from wound (red) and control (orange) skin sites, by steady-state non-MAIT TCRβ+ (dark gray) and by TCRβ− (light gray) cells. Data are representative of two independent experiments (n = 5).
(I) Tissue repair23 signature score on non-cycling MAIT17 cells. Tukey’s multiple comparison test. Please also see Figure S2E.
(J) Average expression of tissue repair23 and MAIT1715 signatures on clusters from (B). Please also see Figure S2F.