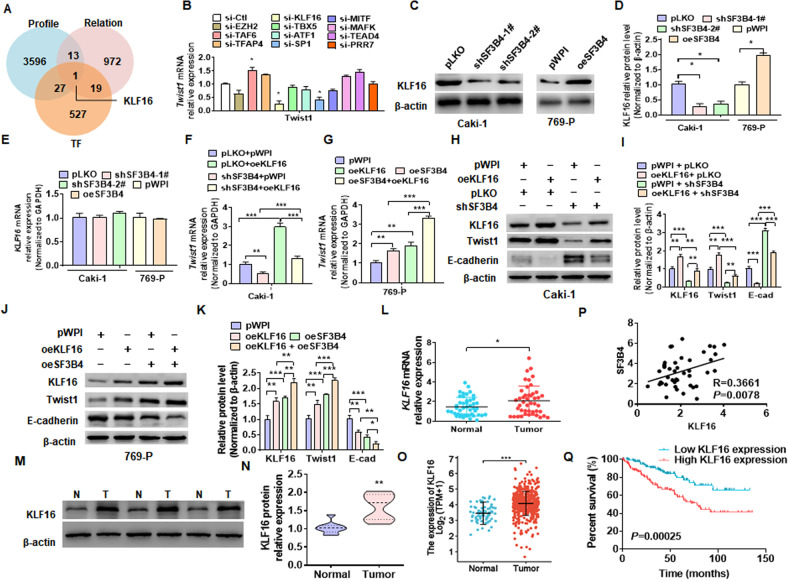
Fig. 5. KLF16 mediates SF3B4 upregulation of Twist1 expression.
A Potential target genes of SF3B4 were analyzed from the data of gene transcriptome of SF3B4-depleted cells (Profile), genes expressed in association with SF3B4 in ccRCC tissues in TCGA data (Relation) and transcription factor (TF), and are shown by a Venn diagram. B RT-qPCR detected the Twist1 mRNA expression in Caki-1 cells transfected with the indicated siRNAs for candidate transcription factors. C, D Western blot examined the KLF16 expression in Caki-1 and 769-P cells transfected with the indicated constructs. E Cells were transfected as in C and then RT-qPCR detected the KLF16 mRNA expression. F, G Caki-1, and 769-P cells were transfected as indicated, and then RT-qPCR detected the expression of Twist1 mRNA. H, I Caki-1 cells were transfected with pWPI-KLF16 (oeKLF16) and shSF3B4, alone or together, and then the protein levels of KLF16, Twist1, and E-cadherin were examined by western blot. J, K 769-P cells were transfected with oeKLF16 and oeSF3B4, alone or together, and then KLF16, Twist1, and E-cadherin were examined by western blot. L KLF16 mRNA expression was detected by RT-qPCR in normal (N, n = 53) or ccRCC (T, n = 53) tissues. M KLF16 protein level was examined by western blot from three pair of randomly selected clinical ccRCC samples. N Quantitative analysis of M. O The expression level of KLF16 was downloaded and then analyzed from data of the TCGA database in normal (n = 72) and ccRCC (n = 533) tissues. P The correlation between SF3B4 and KLF16 mRNA level in ccRCC tissues was analyzed by Pearson correlation analysis of our clinical data (R = 0.3661, P = 0.0078). Q According to the TCGA database, the overall survival for ccRCC patients with low and high Twist1 levels were analyzed by using the Kaplan–Meier method. All data are expressed as the mean ± SEM of three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001 vs. their corresponding controls.