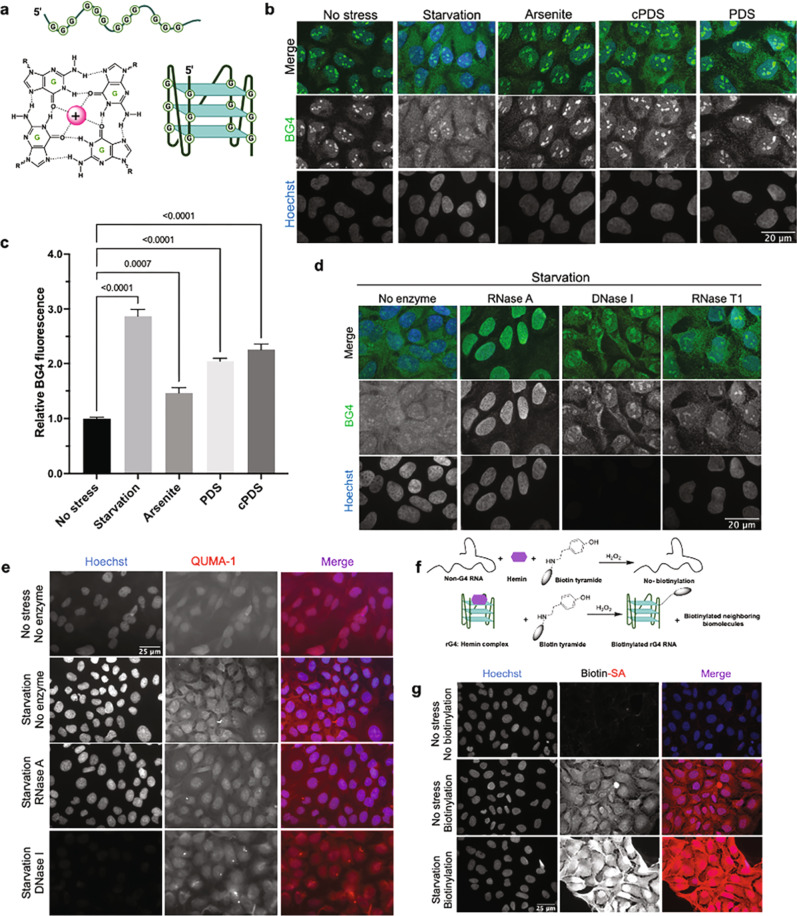
Fig. 1. RNA G-quadruplex formation is enhanced under stress in U2OS cells.
a A square planar G-quartet arrangement stabilized by a monovalent cation (H-bonds and cation-oxygen coordination is shown with dotted bonds), and a schematic of 3-tier G-quadruplex structure stabilized by monovalent cations. The schematic depicts 3 G-quartets (light green planes) stacking with RNA-backbone in a parallel orientation. b BG4 staining in U2OS cells demonstrated both RNA and DNA can form G4s in U2OS cells, and rG4 folding (cytosolic BG4 fluorescence) in the U2OS cells is increased under various stress conditions as indicated by more intense fluorescence signals of BG4 antibody under 100 µM sodium arsenite mediated oxidative stress and starvation stress, rG4, and G4 stabilizing ligands carboxypyridostatin (cPDS) and pyridostatin (PDS) were used as positive controls. c Quantification of fluorescence intensity of BG4 signals, the bars represent the mean + /- SD of fluorescence intensity from 50 cells for each treatment (biological triplicate), statistical significance was tested by One-way ANOVA using no-stress cells as controls, Multiple comparisons, Dunnet correction, and p-values provided. d RNase A treatment almost completely abolished the cytoplasmic BG4 signaling, while DNase I treatment minimized nuclear BG4 staining, and RNase T1 treatment had little effect on resultant cytosolic BG4 fluorescence suggesting cellular RNA is folded in rG4 under stress. e rG4 specific QUMA-1 fluorescence dye was used as a complementary approach to BG4 staining, fixed cells were treated with 2 µM QUMA-1 for 15 min, and imaged. f, g Hemin-bound rG4s have peroxidase activity which can be tuned to biotinylate the corresponding RNA and potentially its interactome. f Schematic of rG4-hemin-biotin tyramide reaction, and g enhanced biotinylation of starved cells in comparison to the non-stressed cells clearly indicates a higher rG4 folding under stress.