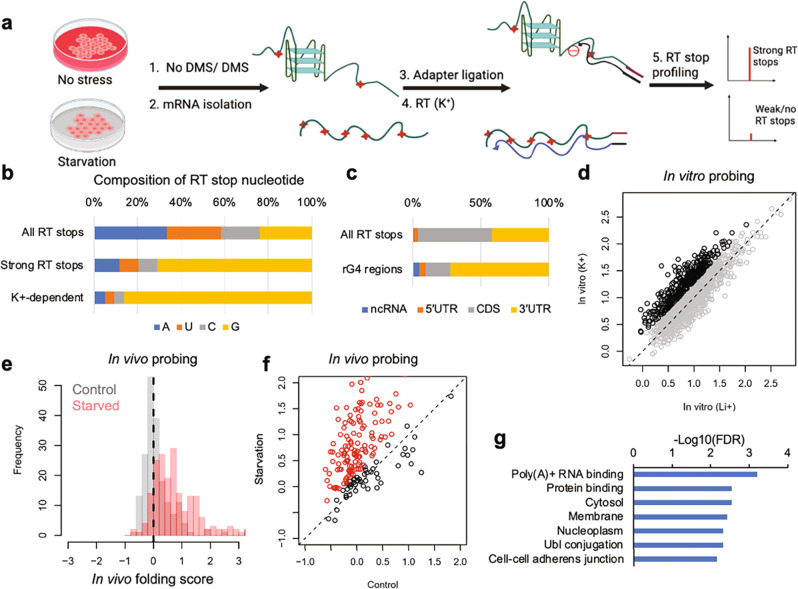
Fig. 2. DMS-mediated unbiased structure probing shows mRNA rG4s are folded under stress in vivo.
a A schematic of DMS-seq and RT-stop profiling to identify the bonafide rG4s in U2OS cells under starvation stress. b RT stops are highly enriched at G residues. c RT stops are selectively enriched in mRNA 3'UTRs. d On average, RT stops in 150 mM K+ folding followed by DMS-modified mRNA are 1.6-fold stronger than that in 150 mM Li+ folding followed by DMS-modified mRNA. rG4 regions showing ≥2 fold difference in DMS accessibility between K+ and Li+ conditions are shown in black. e, f in vivo rG4 folding increases significantly in starved U2OS cells as compared to unstressed cells. g mRNAs containing starvation-induced folded rG4s are enriched in those encoding RNA-binding proteins, ubiquitin ligases, and both cytosolic and membrane proteins.