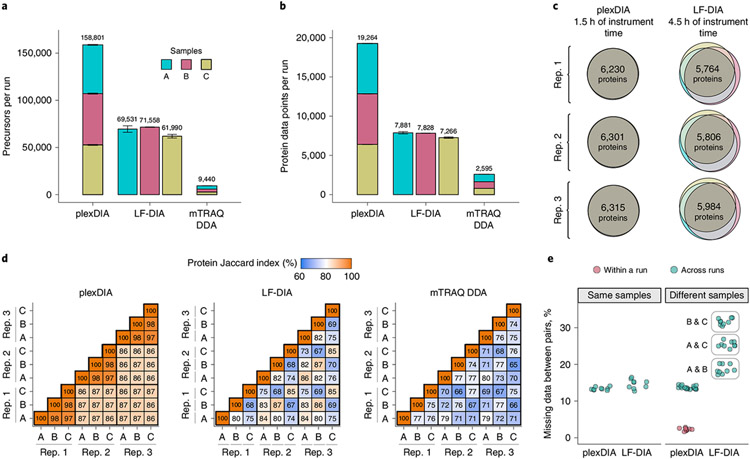
Fig. 2 ∣. plexDIA proteome coverage and overlap between samples and runs.
a, Number of distinct precursors identified from 60-minute active gradient runs of plexDIA, LF-DIA and mTRAQ DDA at 1% FDR. The DIA analysis employed the V1 duty cycle shown in Fig. 1c. Each sample was analyzed in triplicate (n = 3), and the results are displayed as mean; error bars correspond to standard error. b, Total number of protein data points for plexDIA, LF-DIA and mTRAQ DDA at 1% global protein FDR (n = 3). c, Venn diagrams of each replicate for plexDIA and LF-DIA display protein groups quantified across samples A, B and C. The mean number of protein groups intersected across samples A, B and C is 6,282 for plexDIA and 5,851 for LF-DIA. d, The similarity between the quantified proteins across samples is quantified by the corresponding pairwise Jaccard indices to display data completeness. e, Distributions of missing data for protein groups between pairs of runs of either the same sample (that is, replicate injections) or between different samples. All analyses used MBR. The corresponding results for the V2 duty cycle are shown in Extended Data Fig. 3.