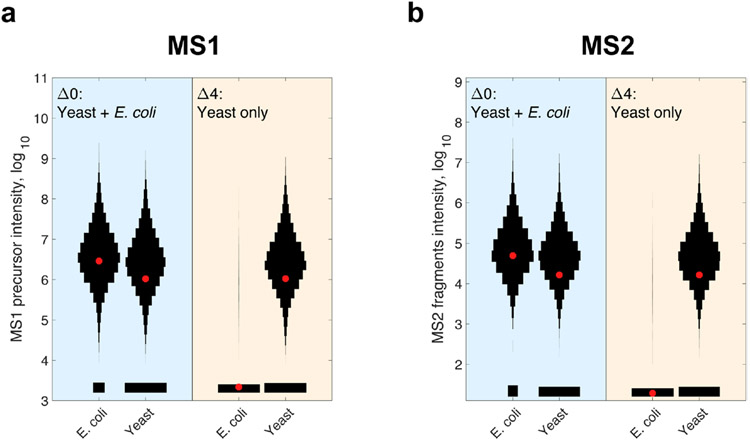
Extended Data Fig. 2 ∣. plexDIA analysis of proteins present only in one samples but missing from another.
We sought to test identification propagation by plexDIA for the case when proteins are present only in some samples and not in others. To do so, we prepared a standard in which one sample (labeled with mTRAQ, 0) had both 0.3 μg E. coli and 0.3 μg S. cerevisiae while another (labeled with mTRAQ Δ4) had only 0.3 μg S. cerevisiae. The combined set was analyzed by plexDIA using the V1 method. (a) Distributions of raw MS1 precursor intensity for E. coli and S. cerevisiae precursors at channel-q-value < 0.01. (b) Distributions of raw MS2 quantification of precursors filtered for channel-q-value < 0.01. The red asterisks correspond to the means of the distributions.