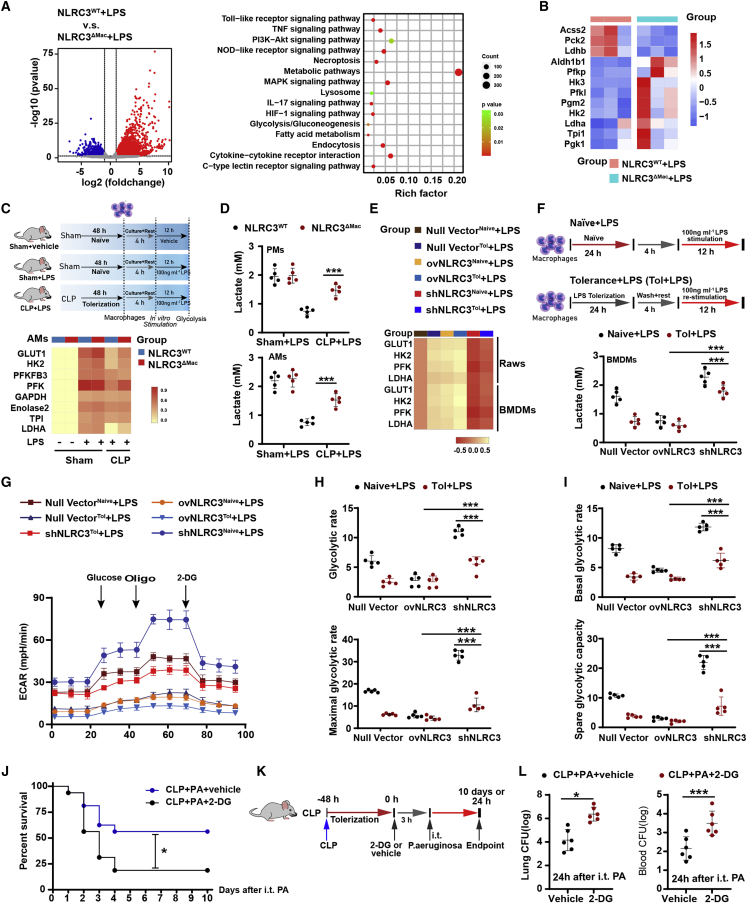
Figure 3.
NLRC3 functions within macrophage to drive glycolysis defects during sepsis-induced immunosuppression
(A and B) Total RNA of each mouse was isolated from 1.0 × 106 BMDMs (n = 3 mice/each group) and analyzed by RNA-seq. (A) Left: scatterplot of the KEGG pathway enrichment analysis of the differentially expressed genes (DEGs) in immunometabolism-associated pathways. Right: the Rich factor is the ratio of DEG numbers annotated in this pathway term to all genes; the size of the symbols represents the number count of DEGs, and higher values indicate greater intensiveness; coloring of the p values indicates the significance of the Rich factor, ranging from −1 to 1. (B) Heatmap showing the relative amounts of RNA of the three pairwise comparisons. (C and E) qPCR detection of the mRNA expression levels of representative NLRC3-regulated genes involved in glycolysis in macrophages: (C) AMs with LPS in vitro challenge after CLP or sham operation (n = 5 mice/each group), and (E) tolerant or naive RAW 264.7 with null vector, stable NLRC3 deficiency (shNLRC3), or overexpression (ovNLRC3) following LPS stimulation. The results of mRNA expression were normalized to β-actin, and log2 values were used to calculate correlations. Each column in the heatmap represents the ratio of normalized expression (n = 5). (D and F) Lactate production in AMs and PMs following LPS in vitro challenge in the indicated genotypes after CLP or sham operation (D) in tolerant or naive BMDMs transduced with shNLRC3 or ovNLRC3 following LPS stimulation (F). (G–I) Analysis of the extracellular acidification rate (ECAR) of tolerant or naive RAW 264.7 macrophages transduced with null or shNLRC3 or ovNLRC3 vector after LPS stimulation. (n = 5). (J–L) Kaplan-Meier survival curves (n = 16) (J); schematic overview of experimental design (K); bacterial loads in the lung (left) and blood (right) from surviving NLRC3ΔMac (LysM-Cre+ NLRC3fl/fl) mice with either 2-DG or PBS after P. aeruginosa intratracheal administration (L). Circles represent individual mice. Graphs show the mean ± SE of experimental replicates and are representative of at least three independent experiments. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; NS, not significant (two-way ANOVA or Student’s t test and log-rank test for survival). See also Figures S7–S13.