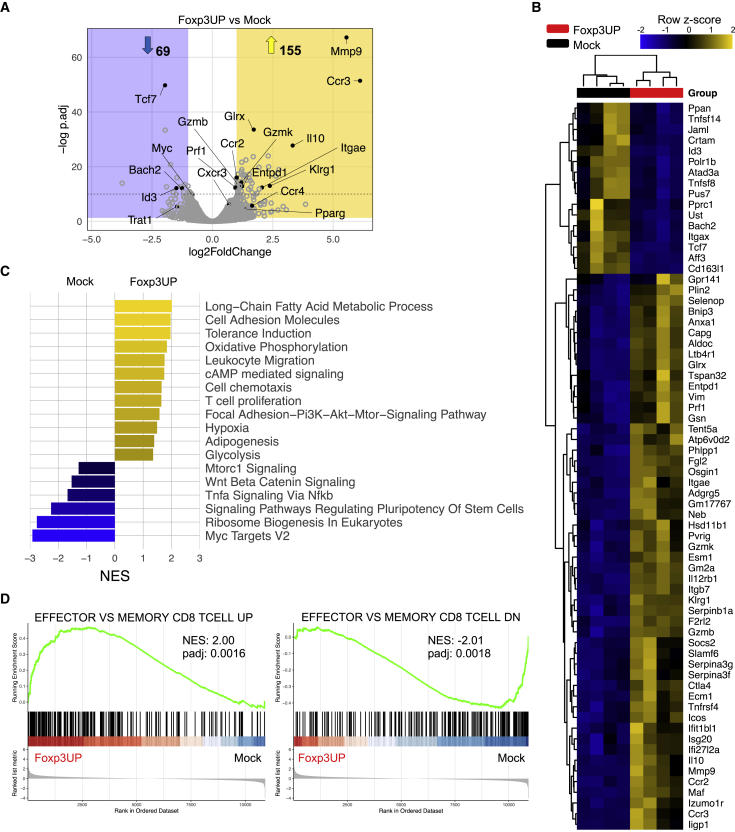
Figure 2.
Transcriptomic signature of Foxp3UP TILs
(A–D) TBI 10-day B16OVA-bearing BL6 (CD45.2+) mice (n = 5) received an i.v. injection of a mix containing 4-day in vitro-expanded mock (CD90.1+) and Foxp3UP (GFP+) (1:1 ratio) OT-I (CD45.1+) cells. Five days later, Foxp3UP (GFP+CD45.1+) and mock (CD90.1+CD45.1+) CD8 T cells infiltrating the tumor were separately isolated by FACS and used for RNA-seq. Differentially expressed gene (DEG) analysis using Foxp3UP and mock CD8 T cells from four independent experiments was performed. (A) Volcano plot depicting DEGs of interest (cutoff log2 fold change > 1, padj < 0.05). (B) Heatmap representation of hierarchical clustering of highly significant DEGs (padj < 10−10). (C) GSEA by clusterProfiler illustrates gene sets positively (normalized enrichment score [NES] > 0) or negatively (NES < 0) enriched in Foxp3UP versus mock TILs. All depicted pathways reached the false discovery rate <0.05 cutoff. (D) GSEA enrichment score curve of “Effector versus memory CD8 T cell” upregulated (UP) and downregulated (DN) gene sets in Fox3UP versus mock TILs presented as the NES.