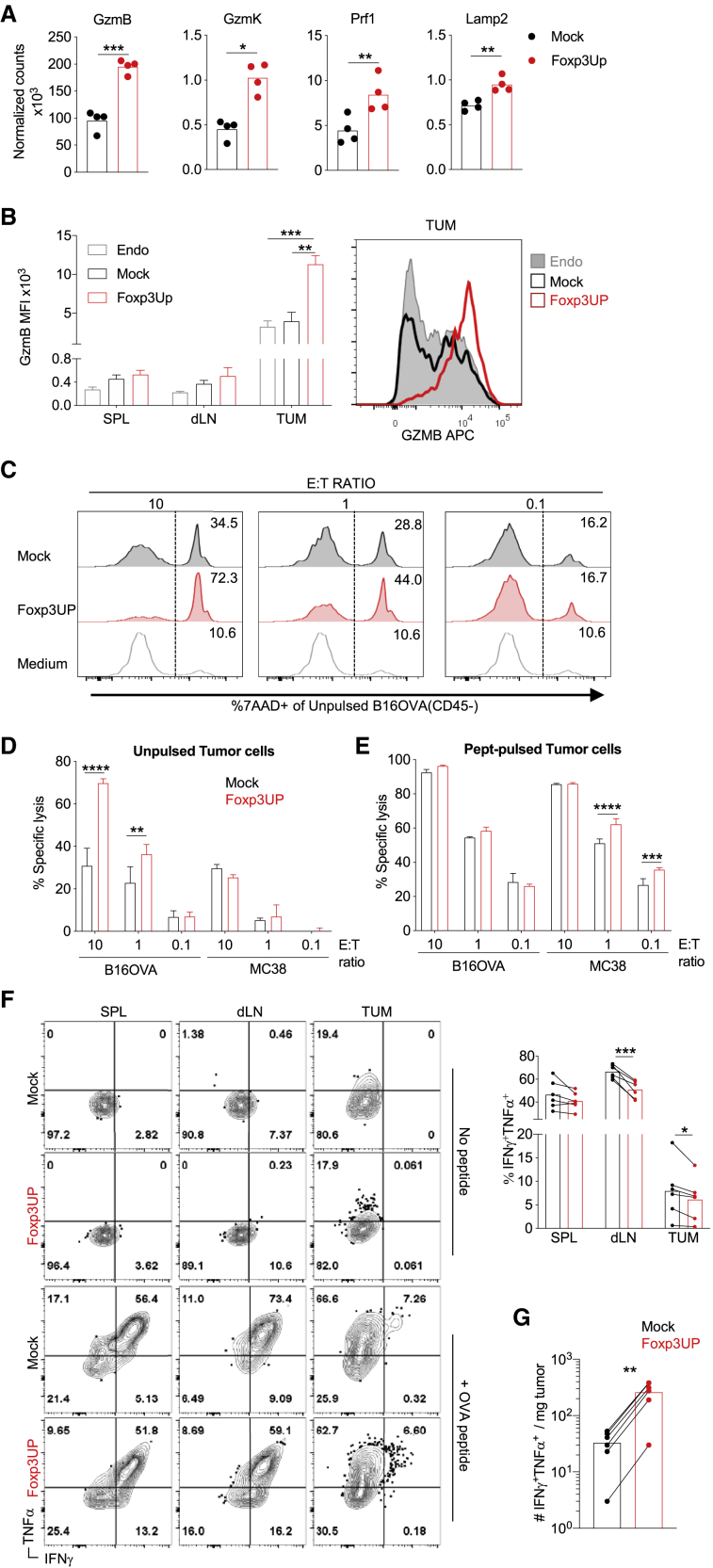
Figure 5.
Enforced FOXP3 expression increased cytotoxicity in CD8 T cells
(A and B) Foxp3UP (GFP+) and mock (CD90.1+) OT-I (CD45.1+) cells were co-injected at a 1:1 ratio as in Figure 2. (A) Normalized counts from an RNA-seq analysis of key cytotoxic genes in Foxp3UP (GFP+CD45.1+) and mock (CD90.1+CD45.1+) CD8 T cells isolated from tumors. (B) The graph on the left shows GzmB median fluorescent intensity (MFI) in endogenous (CD45.2+) CD8 T cells (Endo) and transferred Foxp3UP (GFP+CD45.1+) and mock (CD90.1+CD45.1+) CD8 T cells in the spleen (SPL), dLNs, and tumor (TUM). Histogram on the right depicts GzmB expression in endogenous and transferred TILs. (C–E) mock or Foxp3UP Pmel cells were co-cultured with B16OVA or MC38 tumor cells that had been previously pulsed or not (unpulsed) with Pmel peptide, at different ratios of effector cells to tumor cells (E:T ratio). As a control, tumor cells were cultured alone (medium). The percentage of dead tumor cells (7AAD+CD45−) in total tumor cells (CD45−) was analyzed 12 h later by FACS. (C) Representative histograms showing percentage of dead B16OVA cells at different E:T ratios. (D and E) Killing activity of Pmel cells against unpulsed (D) or pulsed (E) tumor cells depicted as the percentage of specific lysis, as described in materials and methods. (F and G) Mice were treated as in (B). At day 5 of ACT, total cells from the spleen, dLNs, and tumors were restimulated ex vivo with or without OVA peptide, and production of IFNγ and TNFα was assessed 5 h later by FACS. (F) Representative dot plots of cells stimulated with or without peptide (left). The graph on the right shows the percentage of IFNγ+TNFα+ cells within Foxp3UP (GFP+CD45.1+) and mock (CD90.1+CD45.1+) CD8 T cells in spleen, dLNs, and tumor upon peptide restimulation. (G) Number of IFNγ+TNFα+ Foxp3UP and mock OT-I TILs normalized to mg of tumor. Data are presented as mean (A, F, and G), mean + SEM (B), and mean ± SD (D and E). Symbols represent individual mice (F and G) or experiments (A). Statistical significance was determined using unpaired t test (A), paired t test (B, F, and G) and two-way ANOVA for multiple comparisons (D and E). ∗∗∗∗p < 0.00005, ∗∗∗p < 0.0005, ∗∗p < 0.005, ∗p < 0.05. Compiled data from four different experiments (A) or one experiment representative of two experiments (B–G) are shown.