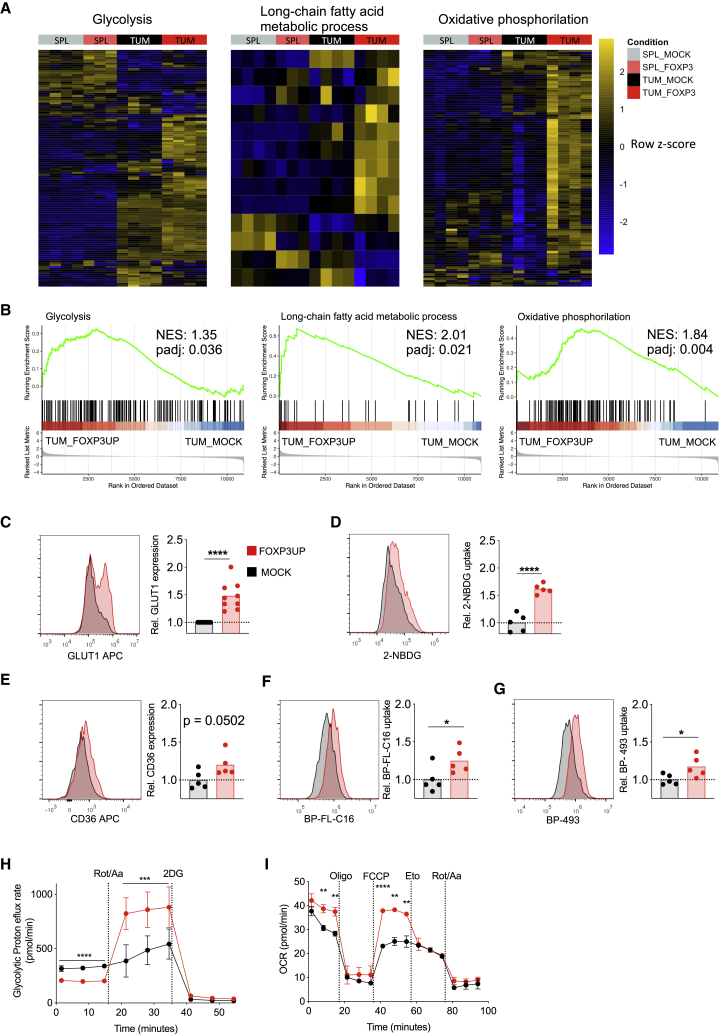
Figure 6.
FOXP3-overexpressing CD8 T cells exhibited improved glycolytic and lipidic metabolism
(A and B) Foxp3UP (GFP+) and mock (CD90.1+) OT-I (CD45.1+) cells were co-injected at 1:1 ratio as described in Figure 2. Transcriptomic profile of Foxp3UP (GFP+CD45.1+) and mock (CD90-1+CD45.1+) CD8 T cells isolated from tumor and the spleen at day 5 of ACT. (A) Heatmap representation of hierarchical clustering of genes differentially expressed (p < 0.05) from hallmark glycolysis, KEGG long-chain FA metabolism, and OXPHOS gene set. (B) GSEA enrichment score curve of glycolysis, long-chain FA metabolism, and OXPHOS pathway in Foxp3UP versus mock TILs presented as the normalized enrichment score (NES). (C–G) Foxp3UP (CD90.1+) and mock (CD90.1+) OT-I (CD45.1+) cells were separately injected into B16OVA tumor-bearing BL6 (CD45.2+) mice, and CD90.1+CD45.1+ TILs were analyzed at day 5 upon ACT. Representative histograms and graphs showing GLUT1 expression (C), ex vivo 2-NBDG uptake (D), CD36 expression (E), ex vivo uptake of palmitate analog Bodipy-FL-C16 (BP-FL-C16) (F), and intracellular lipid droplet staining with Bodipy-493 (BP-493) (G) of Foxp3UP and mock cells. Data are presented as relative MFI values (MFI of studied cells divided by the average MFI of mock CD8 T cells). (H) GlycoPER assay. Before the assay, Foxp3UP and mock CD8 T cells were restimulated in vitro (2 h) with soluble anti-IgG-crosslinked anti-CD3 mAb. GlycoPER was measured at baseline and following injections with rotenone/antimycin A (Rot/AA) and 2-DG. (I) OCR assay under starving conditions. Foxp3UP and mock CD8 T cells were preconditioned (overnight) in substrate-limited growth medium and maintained in poor-nutrient Seahorse medium throughout the Seahorse assay. OCR was measured at baseline and in response to Omy, FCCP, Eto, and Rot/AA. Data are presented as mean (C–G) and mean ± SD (H and I). Symbols represent individual mice (C–G). Statistical significance was determined using unpaired t test (C–I). ∗∗∗∗p < 0.00005, ∗∗∗p < 0.0005, ∗∗p < 0.005, ∗p < 0.05. Compiled data from four different experiments (A and B) or one experiment representative of two (C–G) or three (H and I) experiments are shown.