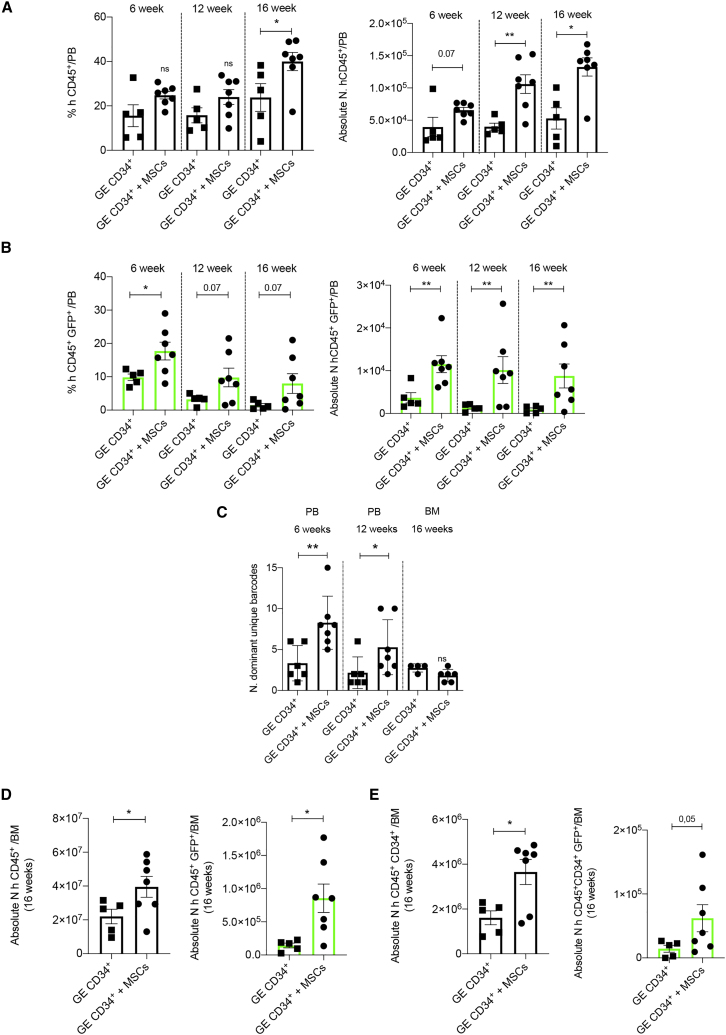
Figure 3.
Improved HSCT outcome after transplantation of HSPCs gene edited in the presence of MSCs into a pre-clinical xenograft model
(A) Flow cytometry analysis of human hematopoietic cell engraftment represented as percentage of hCD45+ cells on total live cells based on physical parameters in the peripheral blood (PB) of mice transplanted with GE-HSPCs co-cultured with BM-MSCs (rounded points) or according to standard protocol (squared points) at different time points after transplantation (left panel). The absolute number of hCD45+ cells was determined by flow cytometry adding known amount of Count Beads to the blood sample and using the counting equation according to the manufacturer protocol of absolute counting beads for flow cytometry (right panel). (B) Engraftment analysis of HDR gene-edited (GFP+) human CD45+ cells on total live cells based on physical parameters in the PB of transplanted mice at different time points after cell infusion (left panel). The absolute number of hCD45+ cells was determined by flow cytometry using absolute counting beads (right panel). (C) Number of unique dominant barcodes identified in the PB (6 and 12 week time points) and in the BM (16 week time point) of mice transplanted with GE-HSPCs + MSCs (GE CD34+ + MSCs) or standard GE-HSPCs (GE CD34+) at different time points after cell infusion. (D) Absolute number of human CD45+ cells (left panel) and human HDR gene-edited (GFP+) CD45+ cells (right panel) on total live cells based on physical parameters engrafted in the BM of transplanted mice at sacrifice. (E) Absolute number of human CD34+ (left panel) and HDR gene-edited (GFP+) CD34+ (right panel) cells within the human CD45+ engraftment on total live cells in the bone marrow at sacrifice (16 weeks). For all plots, individual data points represent an animal. p values were determined by Mann-Whitney test (∗p ≤ 0.05; ∗∗p ≤ 0.001).