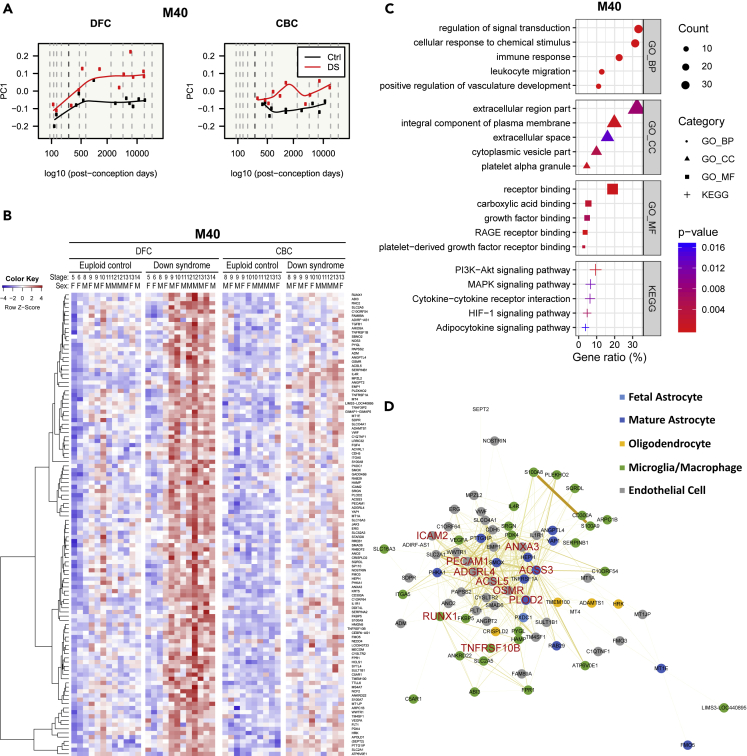
Figure 11.
Characterization of the most DS-associated module
(A–D) Gene expression patterns of the most DS-associated module (M40) are visualized in a line graph of module eigengenes (A) and a gene expression heatmap (B). Functional enrichment analysis with genes related to GO terms and KEGG pathways on M40 (C). The five most significant terms (p< 0.05) are shown in each category. GO: gene ontology, BP: biological process, CC: cellular component, MF: molecular function, KEGG: Kyoto Encyclopedia of Genes and Genomes pathway. Intramodular co-expression networks of M40 (D). Nodes indicate genes, and the width and darkness of edges are proportional to the co-expression level between two genes. The 10 genes with the highest kME values were selected as hub genes and are marked with large red labels. The nodes of genes that showed higher expression in a brain cell type are colored (astrocyte: blue, microglia: green, endothelial cell: grey, and oligodendrocyte: yellow), and the nodes of genes that are not related to any cell type are colored white. Genes of M40 were upregulated in DS and related to cell signaling, immune response, vasculature, and extracellular space.