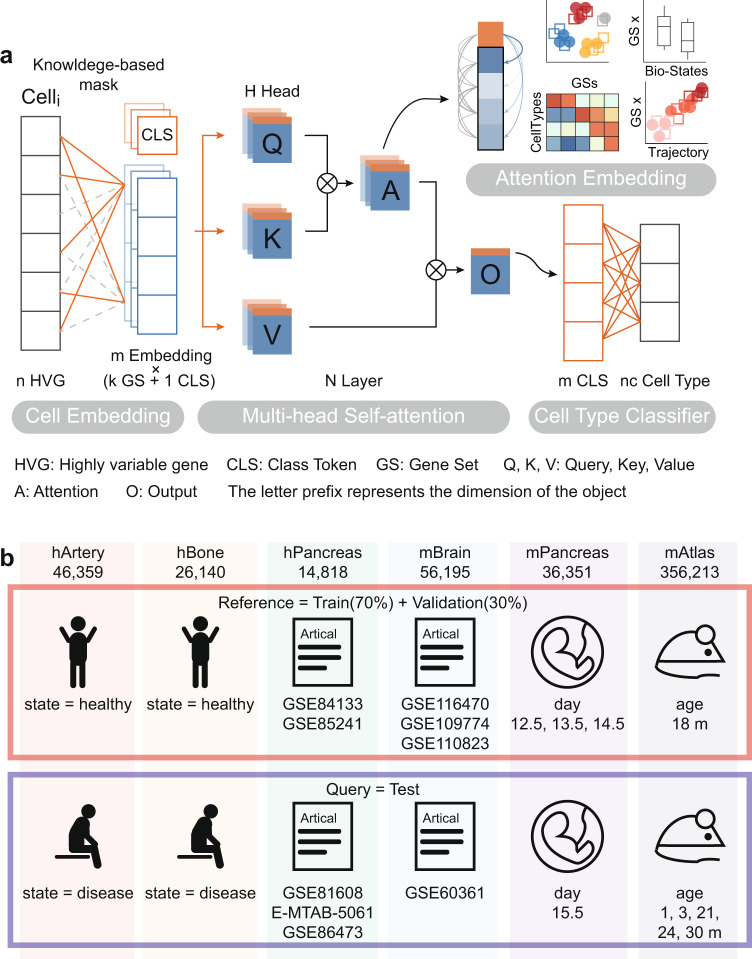
Fig. 1. Algorithmic framework of TOSICA.
a The model is trained on single-cell RNA sequencing data and cell type label for each cell. Based on databases or expert knowledge, masked learnable embeddings are used to convert the reference input data (n genes) to k input tokens representing each gene set (GS), to which class token (CLS) is added. In the attention function, query (Q), key (K), and value (V) matrix are linearly projected from these GSs and CLS combined tokens and the weights (attention, A) is computed by a compatibility function of the Q with the corresponding K, then assigned to each V for computing output (O). In each Multi-head Self-attention layer, the attention function is performed H times in parallel. The CLS of O, considered as latent space of each cell, is used as input of the whole conjunction neural network cell type classifier. Meanwhile, the attention of class (CLS) token to gene set (GS) tokens is referred as attention score and used for cell embedding. b hArtery and hBone datasets use healthy samples as training data and predict disease samples. hPancreas and mBrain datasets are split by data source. Training and test data in mPancreas and mAtlas come from different timepoints.