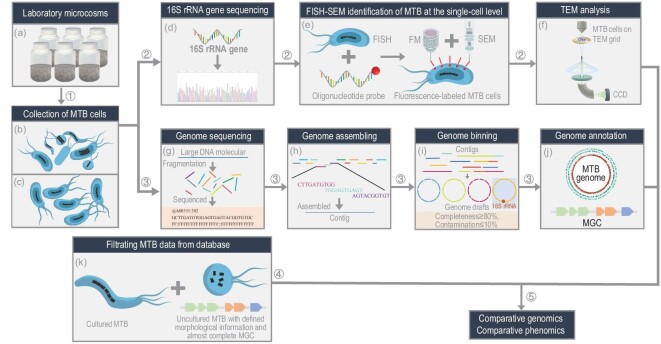
Figure 1.
Five-step workflow for genomic and phenomic study of uncultured MTB. Each step is designed to obtain a specific piece of information. In Step 1, living MTB cells are collected from (a) laboratory microcosms, generally using homemade magnetic separation apparatus or capillary racetrack method. By magnetic separation, living MTB can be collected in sufficient amounts for further morphological and molecular biological studies. Some collections contain different MTB strain types (b), whereas other collections are dominated by one strain type (c). In Step 2, uncultured MTB are identified and characterized at the single-cell level [31], using (d) 16S rRNA gene sequencing of magnetically collected MTB cells, (e) fluorescence in situ hybridization (FISH) of targeted MTB cells with species-specific oligonucleotide probe and coordinated fluorescence microscopy (FM) and scanning electron microscopy (SEM) observations of probe-hybridized MTB cells. This step is generally followed by (f) transmission electron microscope (TEM) analyses, which provide morphological and chemical information on both cells and intracellular magnetic particles down to the atomic scale. Step 3 consists of genomic analyses and generally involves MTB cell (g) genome sequencing, (h) assembling, (i) binning and (j) annotation. Step 4 consists of (k) selecting MTB genome data from public databases. All cultured and uncultured MTB strains with defined morphological information and almost complete magnetosome gene clusters (MGCs) were filtered from the NCBI database. Step 5 consists of integrating genomic and phenomic analyses of cultured and uncultured MTB to understand magnetic particle biogenesis and chain organization within phylogenetically different MTB.