FIGURE 2.
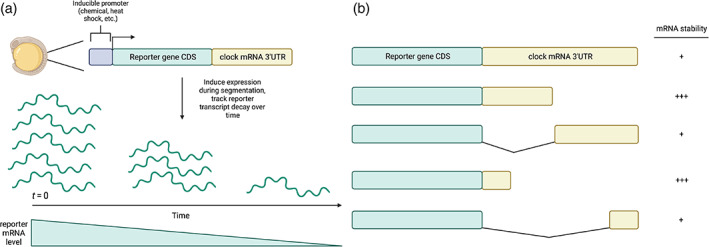
In vivo reporter assays and 3′UTR deletion analysis. (a) Inducible reporter systems and measuring mRNA decay rates. Inducible reporter assays are utilized to measure and compare reporter transcript stability in the context of varied 3′UTR sequences. Depending on the promoter sequence used, these constructs can be chemically or heat shock‐induced in segmenting embryos, when the appropriate segmentation clock transcript regulatory factors are expressed. To calculate reporter transcript decay rates, RNA is extracted from embryos collected at regular intervals post‐induction and reporter mRNA is subsequently quantified across time points using real‐time PCR. (b) 3′UTR fragmentation and reporter transcript decay analysis. Because 3′UTR sequences are rich in motifs that may or may not influence stability, the generation of a set of reporters containing varying portions of a 3′UTR can help to identify smaller regions that influence reporter stability. Upon identification of minimal regions that influence reporter stability, motif analysis followed by mutagenesis of potential regulatory elements can uncover miRNA and/or RBP binding sites that are the primary regulators of mRNA stability. Image created using Biorender.com
