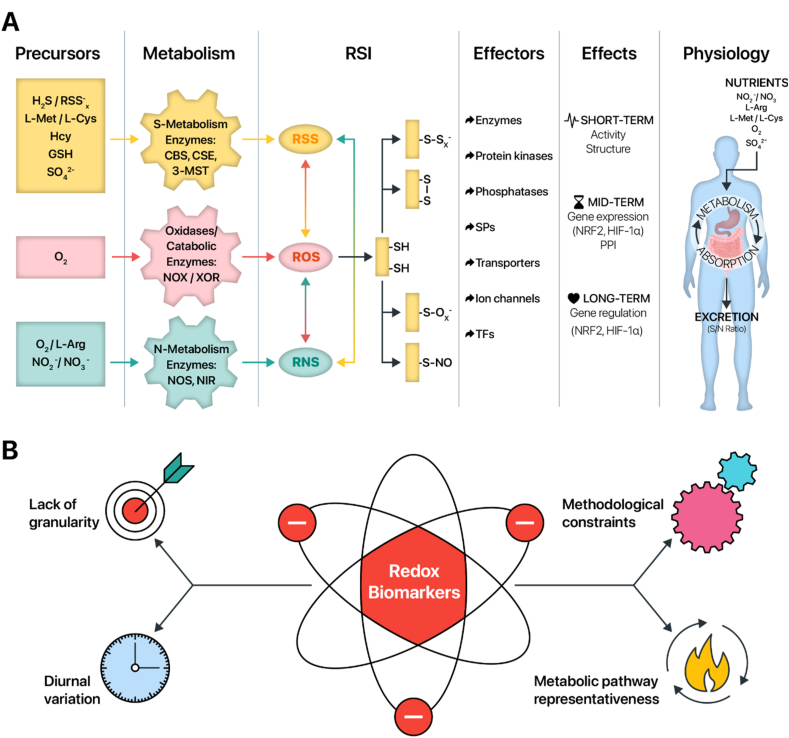
Fig. 1.
A conceptual framework and key considerations to foster the development of redox biomarkers for whole-body redox status in IBD. (A) Schematic overview of the key elements of the Reactive Species Interactome (RSI) which aims to describe chemical interactions among the different types of reactive species (ROS, RNS, and RSS) as well as their interactions with downstream biological targets, e.g. cysteine-based redox switches. Precursors of the RSI are mainly derived from the diet, consisting of organic (e.g. l-methionine, l-arginine) and inorganic (e.g. H2S, O2) compounds and cofactors (e.g. pyridoxal-5′-phosphate), which fuel intermediary metabolism and lead to the controlled formation of reactive species. The main biological targets of the RSI are cysteine-based redox switches, or thiols, acting as multimodal redox relays and modulating the activity of signaling molecules, leading to short-term (e.g. protein structure), mid-term (e.g. gene expression) and longer-term adaptations (e.g. gene regulation). Circulating levels of free thiols and stable end products of the RSI also serve as a communication conduit connecting the intestinal supply of RSI precursors with downstream intracellular thiol targets. (B) Key aspects considering the future of redox biomarkers in IBD, including a lack of granularity of biomarkers, methodological constraints, diurnal variation and their representativeness of redox-related metabolic pathways. Abbreviations: 3-MST, 3-mercaptopyruvate sulfurtransferase; CBS, cystathionine β-synthase; CSE, cystathionine γ-lyase; L-Arg, l-arginine; L-Cys, l-cysteine; L-Met, l-methionine; NIR, nitrite reductase; NOS, NO synthase; NOX, NADPH oxidases; RNS, reactive nitrogen species; ROS, reactive oxygen species; RSI, Reactive Species Interactome; RSS, reactive sulfur species; SPs, structural proteins; TFs, transcription factors; XOR, xanthine oxidoreductase.