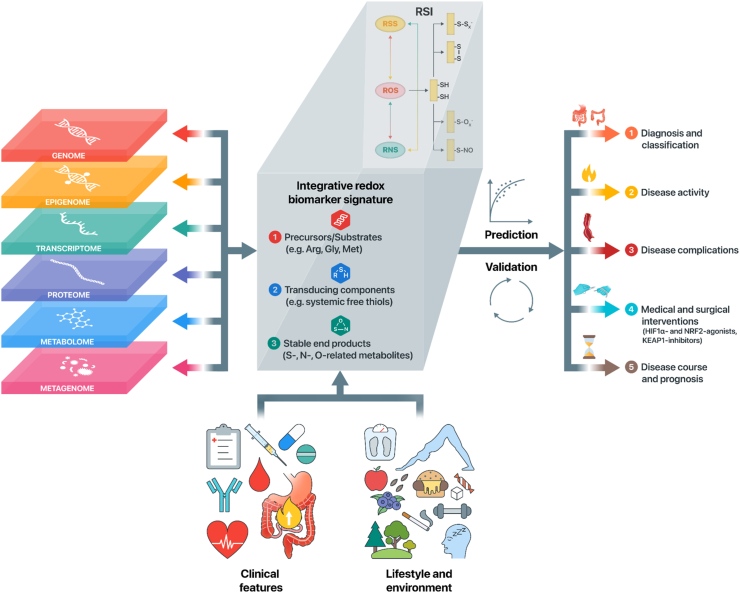
Fig. 3.
Personalized redox medicine and redox metabolomics in the context of IBD. An unbiased, data-driven multi-omics approach may help to define the key components of an integrative redox biomarker signature, while taking into account clinical features and lifestyle-related and environmental factors. Integrative redox-omics, i.e. analyzing redox biomarkers across different levels of biological organization, may help identify the key biomarkers based on the RSI. While integrating all this information, however, it is critical to allow careful phenotypic patient stratification at the same time. After the identification of predictive redox biomarker signatures, validation (both internally and externally) in independent cohorts should be pursued to reliably evaluate their utilities. Ultimately, this may lead to outcome-specific biomarker signatures that may help to (1) aid in diagnosis and improve molecular reclassification of IBD (e.g. disease location or extent), (2) to discriminate between quiescent and active disease states, (3) to discriminate between different types of disease complications (e.g. stricturing and penetrating disease phenotypes), (4) to predict individual responses to medical and surgical interventions in IBD, and (5) to help predict the risk of post-surgical disease course and, eventually, disease prognosis. Abbreviations: Arg, arginine; Gly, glycine; Met, methionine; RSI, Reactive Species Interactome.