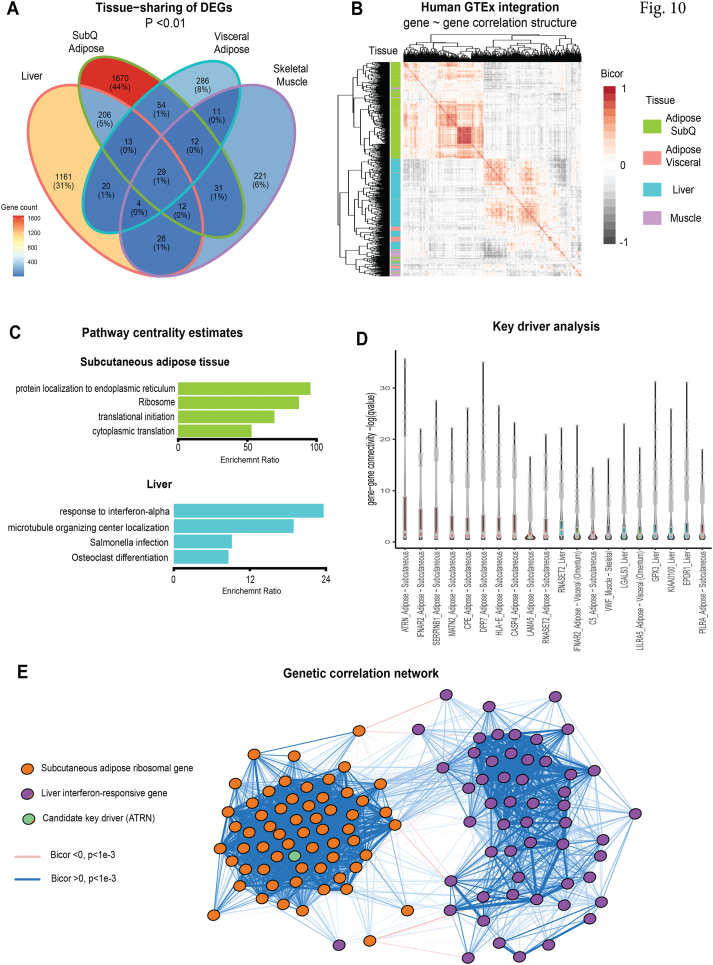
Figure 10.
Integration of mouse differentially expressed genes (DEGs) with human genetic correlation structure. A) Venn diagram showing number of differentially expressed (logistic regression p-value <0.01) genes per tissue analyzed. B) Heatmap showing co-correlation structure of all human orthologues from the Genotype-Tissue Expression (GTEx) encoding mouse DEGs. Filled color indicates biweight midcorrelation (bicor) coefficient and row color along y-axis indicates tissue of origin for each gene. Rows and columns of genes are clustered hierarchically to order by similarities (mclust). C) All genes among 4 tissues indicated were correlated with DEGs and ranked by significance (mean q-value of regression), the top 500 genes were then assayed for pathway enrichments using overrepresentation analyses in Gene Ontology (Biological Processes), KEGG and Reactome. The 4 strongest pathways enriched are shown for either subcutaneous (inguinal) white adipose tissue (top) or liver (bottom). D) Key driver analysis showing top 20 genes encoding secreted proteins ranked by -log10 (p-value of regression coefficient) across all DEG orthologues in GTEx. E) Undirected network showing gene correlation structure of adipose tissue ribosomal and liver interferon-responsive pathway genes, where a key driver (Attractin, ATRN) is shown as a central component. Logistic regression p-values calculated using DESeq2, n = 6 mice per group. Bicor and associated p-values calculated using WGCNA package and q-values using q-value package. GTEx = 310 individuals.