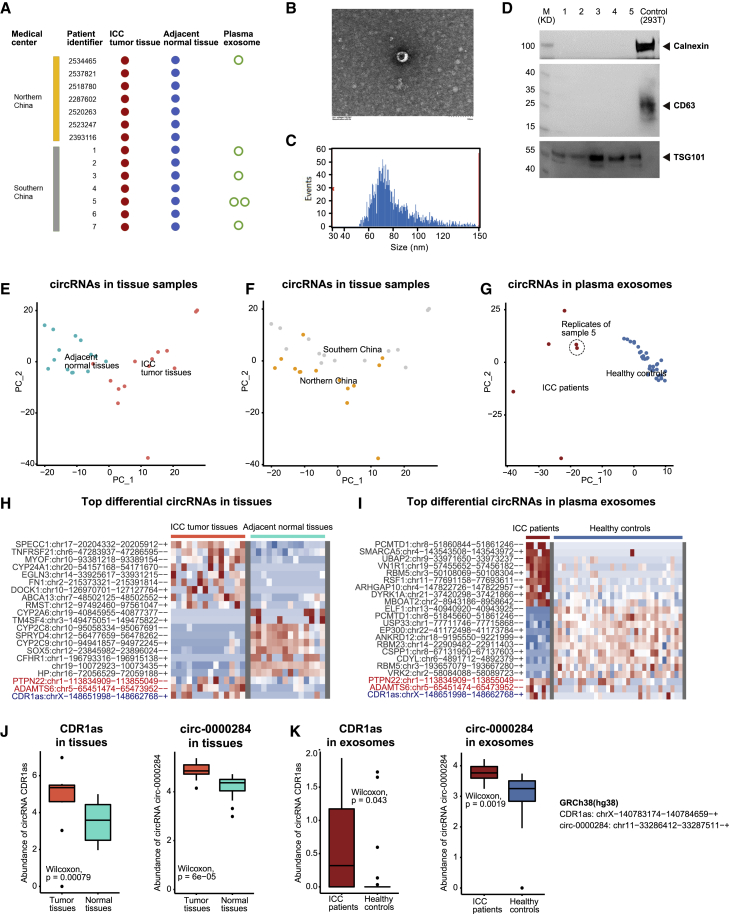
Figure 1.
Landscape of circRNAs in tumor tissues and plasma exosomes of Asian ICC
(A) Sampling and sequencing of the Asian ICC patient cohort. (B) Representative plasma exosomes detected by transmission electron microscopy. (C) The size of plasma exosomes analyzed by high-sensitivity flow cytometry. (D) Western blotting of plasma exosome markers, including TSG101, CD63, and calnexin proteins. (E) Principal-component analysis (PCA) of the circRNA profile in ICC tumor tissues and adjacent normal tissues, labeled by sample types. (F) PCA of the circRNA profile in ICC tumor tissues and adjacent normal tissues, labeled by medical centers. (G) PCA of the circRNA profile in plasma exosomes of patients with ICC and healthy controls (provided by ExoRbase), labeled by sample types. (H) Top differential circRNAs in ICC tumor tissues versus adjacent normal tissues. (I) Top differential circRNAs in plasma exosomes of patients with ICC versus healthy controls. (J) The abundance of CDR1as and circ-0000284 in ICC tumor tissues and adjacent normal tissues. (K) The abundance of CDR1as and circ-0000284 in plasma exosomes of patients with ICC and healthy controls.