Figure 4. Inhibition of GSL biosynthesis decreases the TβRI level in lipid rafts and inhibits the ubiquitination and degradation of TβRI.
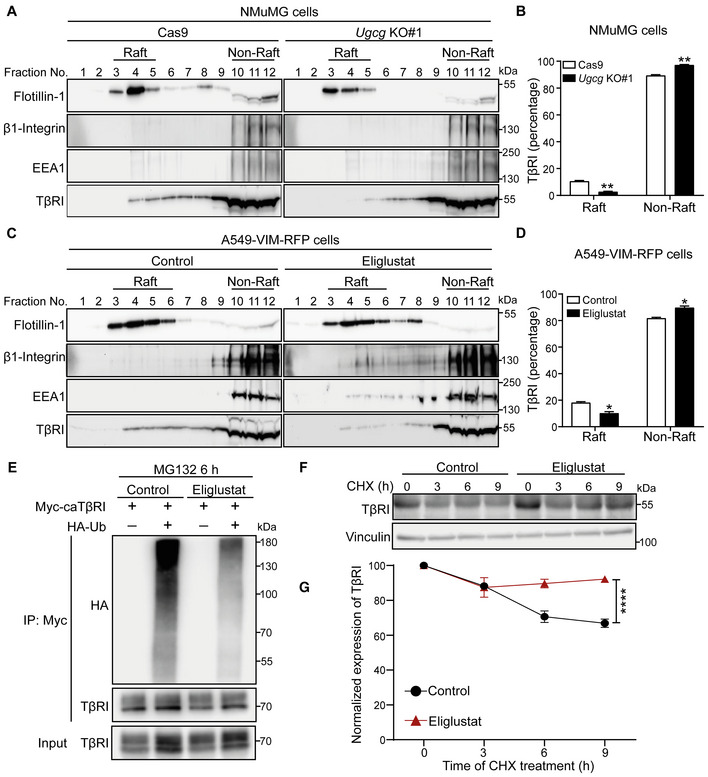
- Lysates from NMuMG control cells (Cas9‐expressing cells) and Ugcg KO cells were subjected to sucrose density gradient ultracentrifugation. The expression levels of flotillin‐1, β1‐integrin, EEA1, and TβRI in the sucrose gradient fractions were analyzed by immunoblotting. Fractions 3, 4, and 5 contained lipid rafts, whereas fractions 10–12 corresponded to the non‐lipid raft fractions.
- Quantification of TβRI percentages in lipid raft and non‐lipid raft fractions from Ugcg KO NMuMG and control cells.
- Isolation of lipid rafts from other cellular components in A549‐VIM‐RFP cells treated with the UGCG inhibitor eliglustat (2 μM) for 6 days using sucrose density gradient ultracentrifugation and measurement of flotillin‐1, β1‐integrin, EEA1, and TβRI levels in the sucrose gradient fractions using immunoblot analysis. Fractions 3–6 contained lipid rafts, whereas fractions 10–12 corresponded to the non‐lipid raft fractions.
- Quantification of TβRI percentages in lipid raft and non‐lipid raft fractions from A549‐VIM‐RFP cells treated with eliglustat (2 μM) for 6 days.
- Ubiquitination of TβRI was detected by immunoprecipitation (IP) of Myc‐tagged caTβRI from HA‐Ub‐transfected HEK293T cells with or without eliglustat (2 μM) treatment for 6 days. All groups were treated with MG132 (5 μM) for 6 h.
- Immunoblot analysis of TβRI expression levels in control and eliglustat (2 μM)‐treated A549‐VIM‐RFP cells treated with 50 μg/ml CHX for the indicated times. Vinculin: loading control.
- Quantification of TβRI expression levels with normalization to the t = 0 controls.
Data information: In (B, D), the data are expressed as the mean ± SD values from two biological replicates (n = 2). In (G), the data are expressed as the means ± SD values from three biological replicates (n = 3). *P ≤ 0.05; **P < 0.01; ****P < 0.0001 based on unpaired Student's t‐test.
