Figure 5. ST3GAL5 inhibits TGF‐β signaling and TGF‐β‐induced EMT.
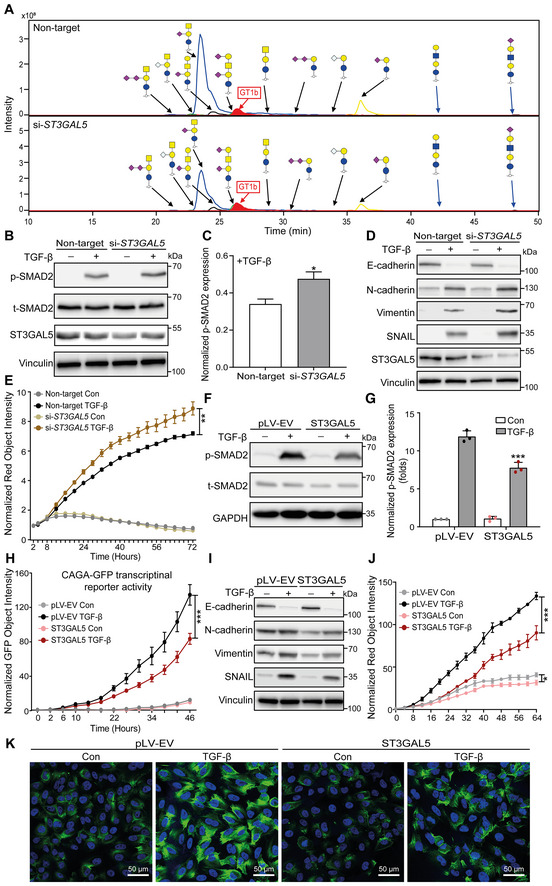
- Extracted ion chromatograms of GSL‐glycans released from A549‐VIM‐RFP cells transfected with nontargeting, ST3GAL5 or B4GALNT1 siRNA. GT1b: internal normalization control.
- Immunoblot analysis of p‐SMAD2, t‐SMAD2, and ST3GAL5 in A549‐VIM‐RFP cells with siRNA‐mediated ST3GAL5 knockdown or transfection of nontargeting siRNA and treated with vehicle control or TGF‐β for 1 h. Vinculin: loading control.
- Quantification of the p‐SMAD2 level in A549‐VIM‐RFP cells transfected with nontargeting or ST3GAL5 siRNA and treated with TGF‐β as shown in (B).
- Expression levels of ST3GAL5, the epithelial marker E‐cadherin, and mesenchymal markers, including N‐cadherin, vimentin, and SNAIL, in siRNA ST3GAL5‐depleted and nontargeting siRNA‐transfected A549‐VIM‐RFP cells treated with vehicle control or TGF‐β for 48 h. Vinculin: loading control.
- The time course of RFP‐tagged vimentin expression was monitored with an IncuCyte system in siRNA ST3GAL5 knockdown and nontargeting siRNA‐transfected A549‐VIM‐RFP cells treated with vehicle control or TGF‐β for the indicated times. The red object intensity was normalized to the red intensity at 0 h.
- Immunoblot analysis of p‐SMAD2 and t‐SMAD2 in A549‐VIM‐RFP cells transduced with empty vector (pLV‐EV) or the ST3GAL5 expression construct and stimulated with vehicle control or TGF‐β for 1 h. GAPDH: loading control.
- Quantification of the p‐SMAD2 level in A549‐VIM‐RFP cells transduced with pLV‐EV or the ST3GAL5 overexpression construct and treated with TGF‐β, as shown in (F).
- A549‐VIM‐RFP cells transduced with the CAGA‐GFP lentiviral vector and with the pLV‐EV control or ST3GAL5 expression construct were treated with vehicle control or TGF‐β for the indicated times. SMAD3/SMAD4‐dependent (CAGA)12‐mediated transcriptional GFP reporter expression levels were monitored with an IncuCyte system. The GFP object intensity was normalized to the green intensity at 0 h.
- Immunoblot analysis of the epithelial marker E‐cadherin and mesenchymal markers, including N‐cadherin, vimentin, and SNAIL, in A549‐VIM‐RFP cells transduced with the pLV‐EV control or ST3GAL5 expression construct and treated with vehicle control or TGF‐β for 48 h. GAPDH: loading control.
- Real‐time expression of RFP‐tagged vimentin was monitored with an IncuCyte system in A549‐VIM‐RFP cells transduced with pLV‐EV or the ST3GAL5 expression construct and treated with vehicle control or TGF‐β for the indicated times. The red object intensity was normalized to the red intensity at 0 h.
- Alexa Fluor 488 phalloidin staining of F‐actin (green) in pLV‐EV control‐ or ST3GAL5‐expressing A549‐VIM‐RFP cells after stimulation with vehicle control or TGF‐β for 48 h. Nuclei were counterstained with DAPI (blue). Images were acquired with confocal microscopy. Scale bar = 50 μm.
Data information: TGF‐β was applied at a final concentration of 2.5 ng/ml. In (C, E, G, H, J), the data are expressed as the mean ± SD values from three biological replicates (n = 3). *P ≤ 0.05; **P < 0.01; ***P < 0.001. In (C), statistical analysis was based on unpaired Student's t‐test. In (E, G, H, J), statistical analysis was based on two‐way ANOVA.
