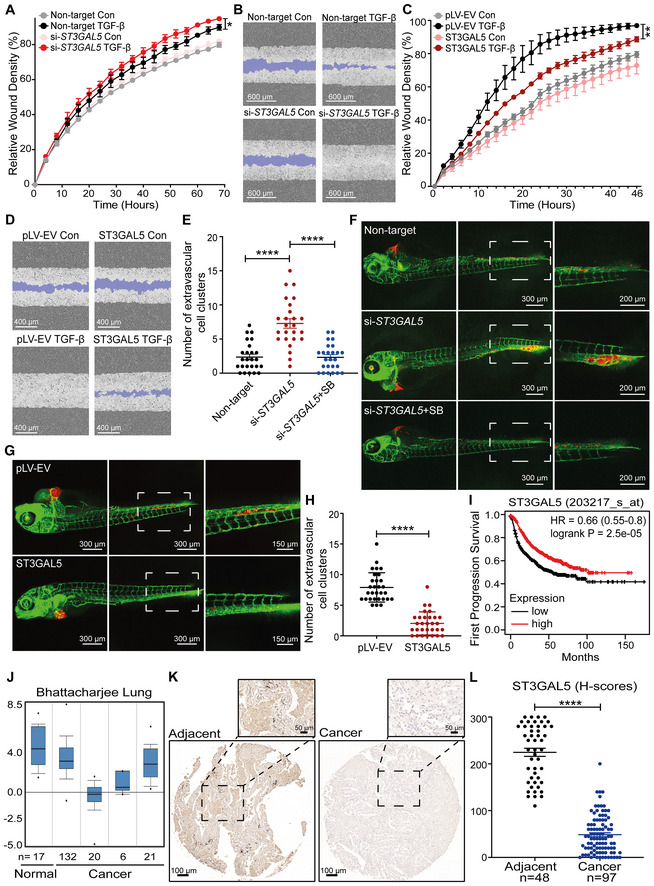
Real‐time scratch assay results were analyzed by an IncuCyte system in ST3GAL5 siRNA‐depleted or nontargeting siRNA‐transfected A549‐VIM‐RFP cells treated with vehicle control or TGF‐β (2.5 ng/ml) for the indicated times. The relative wound density (closure) values are presented as the means ± SD values from three biological replicates (n = 3). *P ≤ 0.05 based on two‐way ANOVA.
Representative images of a scratch wound at the final time point (68 h) in nontargeting siRNA‐transfected and ST3GAL5 siRNA knockdown A549‐VIM‐RFP cells treated with vehicle control or TGF‐β (2.5 ng/ml). The region of the original scratch is indicated in purple. Scale bar = 600 μm.
The time course of wound closure was analyzed by an IncuCyte system in A549‐VIM‐RFP cells transduced with the pLV‐EV control or ST3GAL5 expression construct and treated with vehicle control or TGF‐β (2.5 ng/ml) for the indicated times. The relative wound density (closure) values are presented as the means ± SD values from three biological replicates (n = 3). **P < 0.01 based on two‐way ANOVA.
Representative images of a scratch wound at the 46 h time point in A549‐VIM‐RFP cells transduced with the pLV‐EV control or ST3GAL5 ectopic expression construct. The original scratch region is indicated in purple. Scale bar = 400 μm.
mCherry‐labeled A549 cells with ST3GAL5 siRNA depletion or transfection of nontargeting control siRNA were injected into ducts of Cuvier of zebrafish embryos. Zebrafish embryos in the SB group were treated with the inhibitor SB505124 (1 μM) in the egg water for 4 days after injection with daily refreshment of the treatment. The number of extravasated cell clusters was quantified in 25 embryos per group. ****P < 0.0001 based on unpaired Student's t‐test, n = 2.
Representative images with magnified regions (outlined with dotted squares) of extravasated A549 cells in the indicated groups were acquired 4 days after injection by confocal microscopy. Scale bar = 300 or 200 μm.
In vivo zebrafish extravasation experiments with mCherry‐labeled A549 cells with or without ectopic expression of ST3GAL5. Representative images with magnified regions (outlined with dotted squares) of extravasated cells were acquired 4 days after injection by confocal microscopy. Scale bar = 300 or 150 μm.
The number of extravasated cell clusters was quantified in 30 embryos injected with A549 cells transduced with the pLV‐EV control or ST3GAL5 expression construct. ****P < 0.0001 based on unpaired Student's t‐test, n = 2.
Kaplan–Meier survival curves showing the first progression survival of lung cancer patients in a publicly available lung cancer dataset according to ST3GAL5 expression (n = 982).
Box plots of ST3GAL5 gene expression levels in lung cancer tissues and normal tissues in the Bhattacharjee Lung database. The central bands indicate the medium expression values of ST3GAL5, boxes indicate the expression level ranges of ST3GAL5. Data are presented as the means ± SDs from the indicated number of tissues.
Representative images of ST3GAL5 immunohistochemistry in a human lung tissue microarray including normal and cancer tissues. Wide field and magnified images (outlined with dotted squares) are shown. Scale bar = 100 μm.
Scatter plot showing the expression of ST3GAL5 in normal and cancerous lung tissues. Each point represents the H‐score of a single tissue sample with ST3GAL5 staining ranging from completely absent (H‐score 0) to very strong (H‐score 300). H‐scores are presented as the means ± SDs; normal tissues, n = 48; cancer tissues, n = 97; ****P < 0.0001 based on unpaired Student's t‐test.