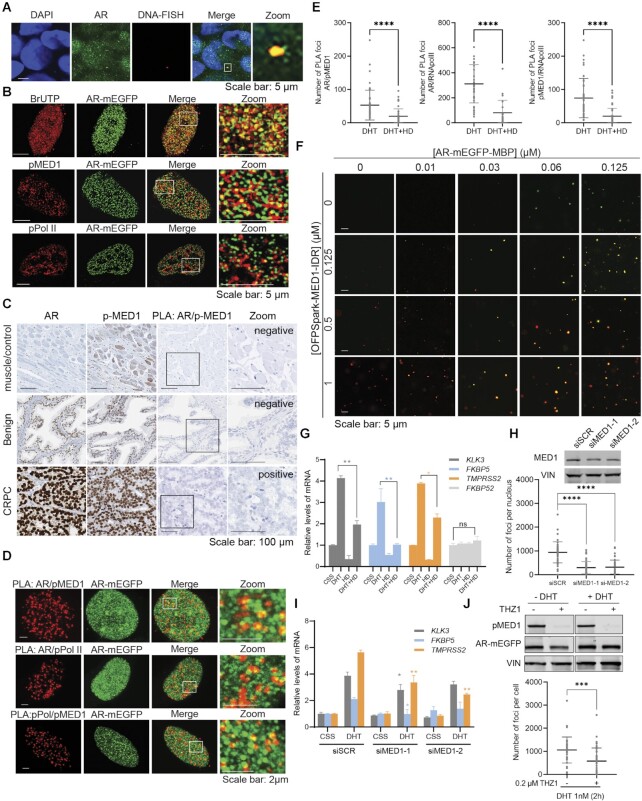
Figure 5.
Foci formation correlates with AR transcriptional activity in mEGFP expressing LNCaP cells. (A) Co-localization between AR foci and the locus of FKBP5 as detected by immunofluorescence (IF) and DNA-FISH in LNCaP cells transfected with AR-mEGFP, starved for 2 days then stimulated with 1 nM DHT for 2 h. The merge between DAPI staining (blue), AR foci (green) and FKBP5-FISH signal (red) is shown. The area of colocalization is enlarged in the zoom panel. (B) IIF imaging of BrUTP, pMED1 and pPol II. LNCaP cells transfected with AR-mEGFP were starved with 5% CSS for two days and then stimulated with 1 nM DHT for 2 h. For BrUTP incorporation assay (top panel), cells were incubated with BrUTP transfection solution for 30 min and then incubated with culture medium without BrUTP for another 2 h before fixation. IF on BrUTP, pMED1 and pPol II were performed (red panels) and the co-localization with AR-rich foci (green panel) were examined under confocal microscope. Images in the white frames were enlarged and displayed in the right panel. (C) The expression levels of AR and pMED1 were assessed in clinical prostate tumor specimens with the Ventana DISCOVERY Ultra autostainer. The interaction between the two proteins was evaluated by proximity ligation assay (PLA). (D) Colocalization of combined PLA staining (red) and AR-mEGFP (green) in LNCaP cells starved in 5% CSS for 2 days then stimulated with 1 nM DHT. Images in the white frames were enlarged and displayed in the right panels. (E) AR-mEGFP transfected LNCaP cells were grown in 5% CSS for two days and then received 1 nM DHT for 2 h. Cells were then treated with or without 4% 1,6-hexanediol (HD) for 5 min before fixing and PLA staining. The quantification of PLA signal was performed using the same method as foci quantification. The aligned points present the data from 45 cells and from three independent experiments with mean ± SD. (F) AR-mEGFP-MBP droplet and MED1-IDR colocalization in vitro. (G) Effect of HD on mRNA levels of different genes. Cells were starved for 3 days and then treated with 1 nM DHT ± 2.5% HD for 30 min, and then washed and incubated with DHT containing medium for 16 h. Total RNAs were extracted and the mRNA levels of genes of interest were examined using qRT-PCR. Values are expressed as mean ± SD. (H-I) Effects of knocking down of MED1 on AR condensates (H) and on AR transactivation (I). (H) LNCaP cells were transfected with AR-mEGFP plasmid and 6 h later with siRNA targeting MED1 (siMED1) or control (siSCR). Cells were then grown in 5% CSS for 2 days and stimulated with 1 nM DHT for 2 h. Cells were fixed and the foci formation was quantified. The reduction of MED1 protein levels was validated by western blot. Vinculin (VIN) was used as a loading control. The aligned points represent data from 45 cells from three independent experiments. (I) Cells transfected with siSCR or siMED1 were grown in 5% CSS media for 3 days and stimulated with 1 nM DHT for 16 h. Levels of AR-targeting genes were examined with q-RT-PCR. The PCR values are expressed as mean ± SD. (J) Inhibiting MED1 phosphorylation by THZ1 (0.2 μM for 2 h) reduces foci formation without affecting AR protein levels. LNCaP cells transfected with AR-mEGFP plasmid were cultured in 5% CSS for 2 days and then treated with or without 0.2 μM THZ1 for 2 h before the stimulation with 1 nM DHT for 2 h. MED1 phosphorylation and AR-mEGFP protein levels were investigated with WB. The number of Foci formation per nucleus was quantified and presented with aligned dotplot as mean ± SD from 45 cells and from three independent experiments. p values are indicated by stars: ns ≥ 0.05, * 0.01 to 0.05, ** 0.001 to 0.01, *** 0.0001 to 0.001, **** < 0.0001.