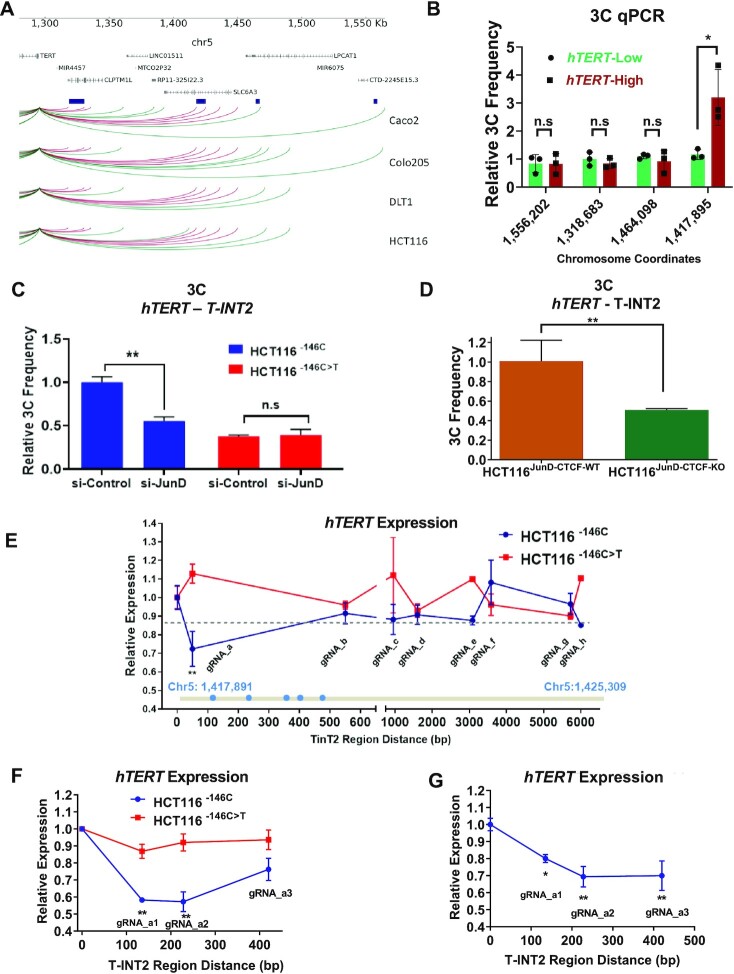
Figure 3.
JunD-mediated CTCF occupancy regulates WT-hTERT–T-INT2 long-range chromatin interaction. (A) 4C sequencing for long-range chromatin interactions with the hTERT promoter in four CRC cell lines. Significant interaction regions (P < 0.05) identified in each line are plotted and regions present in all lines are highlighted in red. Interaction regions are shown in dark-blue boxes, and were functionally validated by 3C-qPCR and CRISPR deletion experiments. (B) Relative chromatin interaction frequency between the hTERT promoter and the indicated chromatin regions identified by 4C were measured by 3C-qPCR in primary CRC samples. Mut-hTERT-specific chromatin interaction region T-INT1 (chr5:1,558,087) was used as a negative control. (C) Chromatin interaction frequency between the hTERT promoter and T-INT2 region was measured by 3C-qPCR in isogenic HCT116 cells transfected with si-Control or si-JunD. (D) Chromatin interaction frequency between the WT-hTERT promoter and T-INT2 was measured by 3C-qPCR in HCT116JunD-CTCF-WT and HCT116JunD-CTCF-KO cells. (E) Isogenic HCT116−146C and HCT116−146C>T cells were co-transfected with KRAB–dCas9 expression vector and different gRNAs targeting the T-INT2 region, or non-targeting control gRNA. RNA was isolated 2 days after transfection and gene expression levels of hTERT were measured by qPCR. The horizontal dotted line shows the mean hTERT expression change in HCT116−146C>T cells and is set as a threshold. The T-INT2 region (chr5:1 417 891–1 425 309) is shown in grey. Putative Sp1 motifs are shown as blue dots within the Sp1 cluster. (F) HCT116−146C and HCT116−146C>T cells were co-transfected with KRAB–dCas9 expression vector and non-targeting control gRNA (cont) or one of three different gRNAs targeting the region targeted by gRNA_a. Gene expression levels of hTERT were measured by qPCR. (G) HCT116–146C cells were co-transfected with dCas9, non-targeting control gRNA (cont) or three different gRNAs targeting the region nearby to gRNA_a. RNA was isolated two days after transfection and gene expression levels of hTERT were measured by qPCR. Ct values were normalized to actin. Error bars indicate mean ± SD of three independent experiments. P-values were calculated by Student's t-test (*P < 0.05; **P < 0.01).