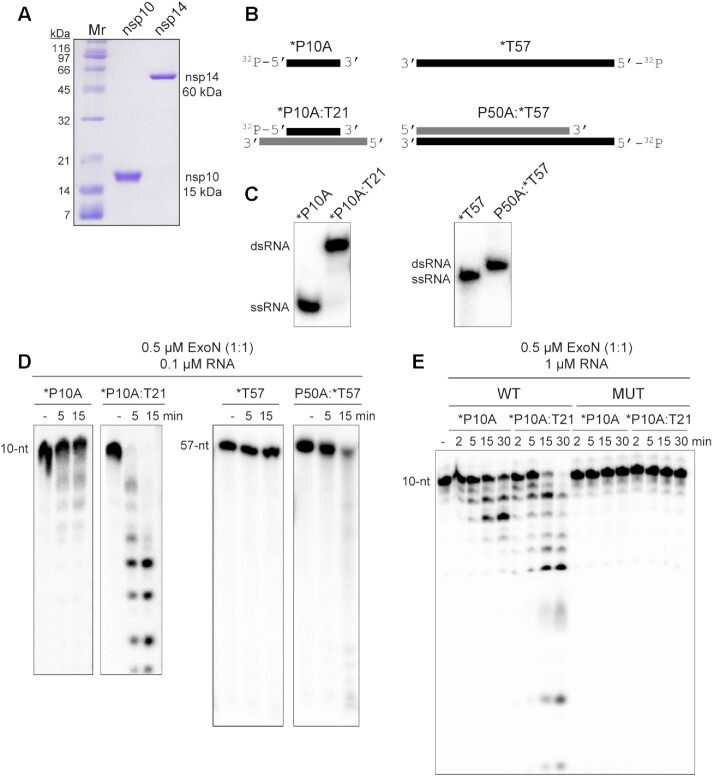
Figure 1.
ExoN prefers primed-template-like dsRNA substrates. (A) Purified SARS-CoV-2 nsp14 (60 kDa) and nsp10 (15 kDa) proteins used in this study. Proteins (1 μg) were resolved on a 15% polyacrylamide gel containing SDS and stained with Coomassie. Broad-range molecular weight markers (Mr) and corresponding molecular weights are indicated. (B) Schematic of ssRNA and dsRNA substrates used to measure exoribonuclease activity. RNA sequences are provided in Table 1 and/or Supplementary Figure S2. RNAs were labeled on the 5′-end with 32P, also indicated by an asterisk (*). RNAs are designated as primers (P) or templates (T). The numbers indicate the length of the RNA. When a base is designated, this reflects the 3′-terminal nucleotide. (C) ssRNA and dsRNA substrates evaluated by Native PAGE. (D) Evaluation of ExoN-catalyzed hydrolysis of RNA. Reaction products were resolved by denaturing PAGE and visualized by phosphorimaging. Reactions contained 0.1 μM ExoN (1:1 nsp14:nsp10) and 0.1 μM of the indicated RNA, were incubated at 30°C for the indicated time, then quenched by addition of EDTA. In both cases, primed-template-like dsRNA substrates were cleaved more efficiently than ssRNA substrates. (E) Exoribonuclease activity is dependent on the nsp14 active site. Residues of nsp14 required for catalysis were changed as follows: D90A, E92A; this derivative is referred to as MUT. Reactions contained 0.1 μM ExoN (1:1 nsp14(WT or MUT):nsp10) and 1 μM RNA and were run as described above. MUT did not exhibit any detectable exoribonuclease activity.