Figure 10.
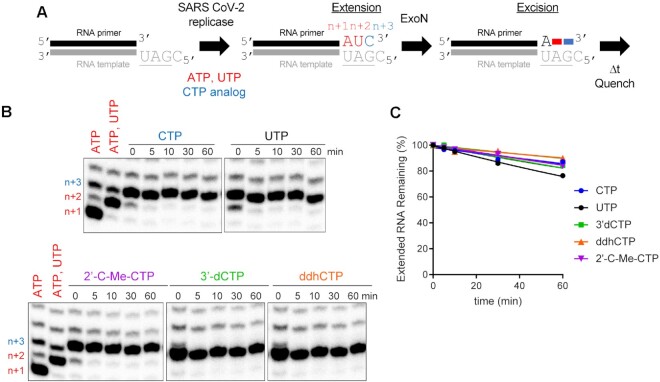
Stalled SARS-CoV-2 replication-transcription complexes block access of ExoN to the RNA terminus. (A) Schematic of assay. Primer extension was initiated by adding the SARS-CoV-2 replicase-transcription complex (nsp7/nsp8/nsp12) in the presence of ATP and UTP or ATP, UTP and CTP or a CTP analog. After an incubation time of 60 min, we added ExoN, incubated for the indicated time, and then quenched the reaction. (B) Effect of ExoN on stalled elongation complexes. Incorporation yields n + 1, n + 2 and n + 3 products. When ATP, UTP and CTP are present, the n + 3 product has a properly basepaired 3′-end. The n + 4 product is formed by misincorporation and has a mispaired 3′-end. When only ATP and UTP (low concentration, 1 μM UTP) are present, both the n + 3 and n + 4 products are formed by misincorporation and both products have mispaired 3′-ends. At high concentrations of UTP (100 μM), most of the products observed are at n + 3 from misincorporation of U opposite G. ExoN was added to elongation complexes for the indicated time (5–60 min). Reaction products were visualized by phosphorimaging after denaturing PAGE. No change in the level of properly paired (n + 3) or mispaired (n + 4) elongation products was apparent for any of the reactions performed, consistent with the replicase remaining bound to both products and obstructing access by ExoN. (C) Quantitation of reaction products shown in panel B. The data were fit to a line. Rates are provided in Table 4.