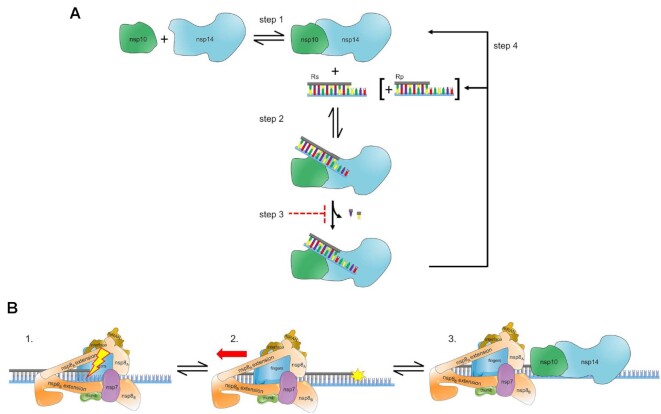
Figure 11.
Hypothetical model for ExoN-catalyzed excision. (A) Excision uncoupled to RNA synthesis. The active exoribonuclease, referred to here as ExoN, is a complex of two subunits: nsp10 (regulatory) and nsp14 (catalytic). The concentration of ExoN therefore depends on the value of the dissociation constant for this binding reaction, as well as the concentrations of nsp10 and nsp14. Most studies to date have ignored this equilibrium. Under steady-state conditions in which the RNA substrate (RS) is present in excess of ExoN, RS binds, catalysis occurs, releasing one or two nucleotides, followed by dissociation of the RNA product (RP). Under these conditions, the rate-limiting step is likely release of RP. Modifications to the terminal nucleotide that reduce the rate of hydrolysis to values substantially lower than the rate of RP dissociation should interfere with excision. RP consumption depends on its ability to compete with RS, for example when more than 20% of the substrate is consumed or when ExoN is present in excess of substrate and more than a single turnover occurs. The possibility also exists for product dissociation to be driven by dissociation of the nsp10 and nsp14 subunits. (B) Excision coupled to RNA synthesis. Introduction of mispaired nucleotides or chain terminators by the polymerase complex does not lead to dissociation, blocking access of the 3′-end for repair by ExoN. The polymerase complex must therefore be actively dislodged from the terminus to access the 3′-end. Once ExoN binds, only one or two nucleotides will be removed.