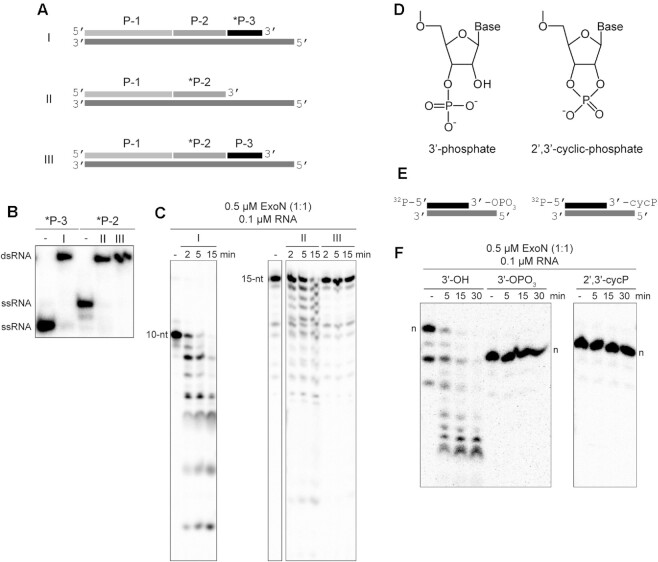
Figure 2.
Products of nsp15-catalyzed endonucleolytic cleavage are not substrates for ExoN. (A) Schematic of dsRNA substrates used. Both substrates I and II contain 32P-labeled RNAs (*P-3 and *P-2) annealed to the template such that both RNAs have an exposed 3′-end. Substrate III contains a 32P-labeled RNA (*P-2) annealed to the template at a position where an additional RNA (P-3) is annealed downstream and blocks access to the 3′-end of the labeled RNA. Substrate III permits assessment of ExoN cleavage at a nick, as produced by nsp15 endonucleolytic cleavage. (B) RNA substrates evaluated by Native PAGE. Shown are the RNAs (*P-2 and *P-3) alone and when annealed to form the dsRNA substrates I, II or III. (C) ExoN does not initiate hydrolysis at a nick. The 10-nt and 15-nt RNAs are indicated. Reactions contained 0.5 μM ExoN (1:1) and 0.1 μM RNA and were quenched at the indicated times. The 32P-labeled RNAs (*P-3 and *P-2) in substrates I and II that have an exposed 3′-end were efficiently cleaved by ExoN. The 32P-labeled RNA (*P-2) in substrate III was not cleaved. (D–F) ExoN does not hydrolyze termini containing a 3′-phosphate or 2′,3′-cyclic phosphate. The structures of these modifications are shown in panel D. Schematic of dsRNA substrates containing 3′-phosphate and 2′,3′-cyclic phosphate modifications used are shown in panel E. *P10A:T21 dsRNA were used as substrates. Reactions contained 0.5 μM ExoN (1:1) and 0.1 μM RNA, were incubated for the indicated time, then quenched. Products are shown in panel F. The unmodified RNA is completely degraded; however, the 3′-phosphate and 2′,3′-cyclic modifications block excision by ExoN. Note, the 3′-phosphate and 2′,3′-cyclic modifications alter the apparent mobility of the RNA as it is more negatively charged and runs faster on the gel; the mobilities of each full-length RNA are indicated by n.