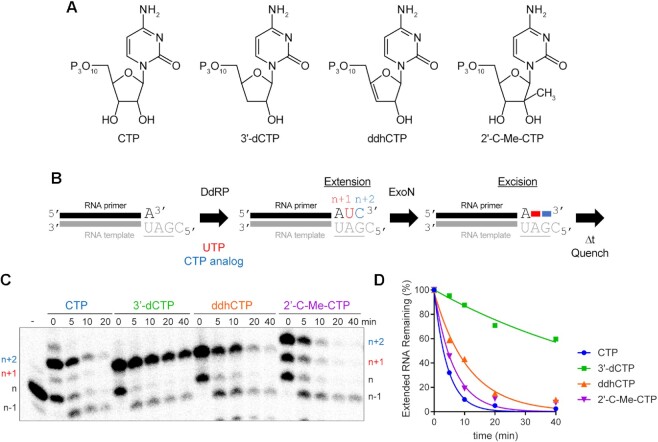
Figure 9.
Viperin product (ddhCMP)-terminated RNA is a good substrate for ExoN but 3′-dCMP-terminated RNA is not. (A) Structure of nucleotide analogs used. (B) Schematic of the assay. Primer extension is initiated by adding an RNA polymerase in the presence of UTP and a CTP analog. Incorporation produces n + 1 and n + 2 products. The last nucleotide to be incorporated is the CTP analog. Once 50–75% of the primers were extended to n + 2, ExoN was added to the reaction. The reaction was monitored over time for hydrolysis. Unextended primer in reactions served as a useful control to demonstrate the presence of active ExoN in the reaction. (C) Analysis of reaction products by denaturing PAGE. Only the 3′-dCMP-terminated RNA exhibited a delay in excision. (D) Kinetics of excision of CMP analogs. The quantitation revealed that only 3′-dCMP-terminated RNA exhibited a significant delay in the rate of excision relative to the CMP control, even though ddhCMP also lacks a 3′-OH. Data were fit to a single exponential. Rates are provided in Table 4.