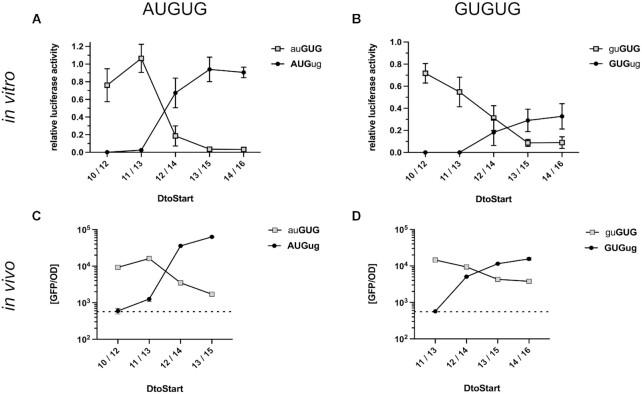
Figure 2.
Open reading frame selection at AUGUG and GUGUG initiation sites. The reporter constructs carried a constant SD sequence (UAAGGAGG) separated from the AUGUG (A, C) or GUGUG (B, D) start codon overlaps by varying spacer lengths. The CDSs of the in vitro luciferase (A, B) and in vivo GFP (C, D) reporter genes were positioned in frame of either the 5′ or 3′ start codon. Expression levels depending on initiation at the 5′ start codon (black circle) and 3′ start codon (grey square) are shown as a function of the spacer length and the corresponding DtoStart. The aligned spacing was varied from DtoStart 10/12 (5′/3′ start codon) to 14/16. GFP reporter constructs harbored the same SD sequence and A-rich spacer elements as the in vitro luciferase reporters. Measured luciferase activities were related to the activity obtained from a reference construct with a distinct AUG start site (see Figure 1A). GFP fluorescence measurements were normalized by optical density (GFP/OD700) and are depicted on a logarithmic scale. Cellular autofluorescent background of an uninduced sample is indicated by a dotted line. For both assays, the mean and the standard deviation of three independent measurements are shown.