Figure 4.
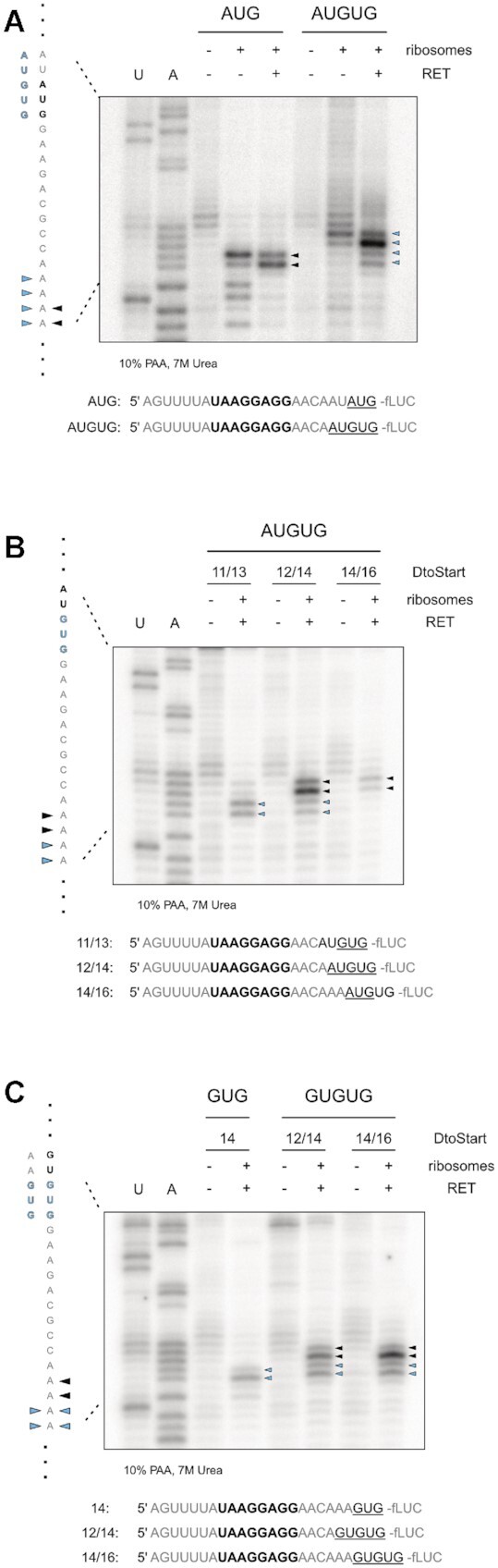
Retapamulin (RET) assisted toeprinting to determine start codon selection at AUGUG and GUGUG initiation sites. (A) Toeprinting analysis of fLUC reporter constructs bearing distinct (AUG) and ambiguous (AUGUG) start sites in the absence and presence of RET. Ribosome specific toeprints, at positions +16 and +17 of the CDS for mRNAs harboring a distinct AUG start codon are indicated with black arrows. At AUGUG initiation sites, toeprint signals representing two populations of initiation complexes are indicated by blue arrows. (B) Toeprinting analysis of start codon selection at AUGUG start sites in dependence of the positioning by a strong SD sequence. Toeprints were performed on the fLUC reporter sequences with aligned spacing of DtoStart 11/13, 12/14 and 14/16. Initiation complexes stalled at the AUG and GUG start codon are indicated by black and blue arrows, respectively. (C) Toeprinting analysis of start codon selection at GUGUG start sites in analogy to (B). Toeprint bands at positions +16 and +17 corresponding to the 5′ GUG start codon are indicated in black, those corresponding to the 3′ GUG in blue. Bands from the distinct GUG control, positioned as the 3′ GUG in the GUGUG sites, are indicated in blue as well. For all depicted toeprint experiments (A–C), sequencing reactions for uracil (U) and adenine (A) bases are shown with the corresponding sequence. The exact RBS context of each mRNA is provided below the respective autoradiograms. The SD sequence is indicated in bold and the selected start codons according to the toeprint patterns are underlined.
