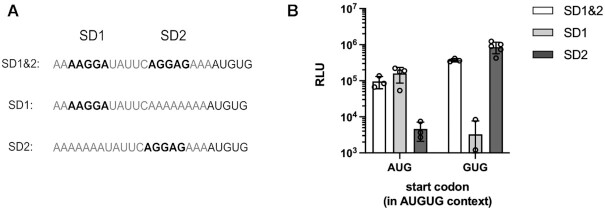
Figure 6.
Mutational study of the tamA RBS to analyze the contribution of the two SD motifs to start codon selection. 20 nts of the native tamA RBS and 15 nts of the corresponding CDS were fused N-terminally to the luciferase reporter sequence. SD mutants were derived from the wild type sequence containing the ambiguous start site and both SD motifs (SD1 & SD2) by substituting SD1 or SD2 with a stretch of As. Replacing the start codon proximal SD motif provided the ‘SD1’ construct, replacing the upstream SD provided ‘SD2’. The tested mutant sequences of the tamA RBS are shown in (A). For each RBS variant, constructs were generated, where the luciferase CDS was placed in frame of either potential start codon. (B) Relative light units (RLU) measured from constructs with luciferase in frame of either AUG or GUG are displayed on a logarithmic scale for the respective RBS sequence variants. The mean and the standard deviation of the independent measurements are shown.