Figure 1.
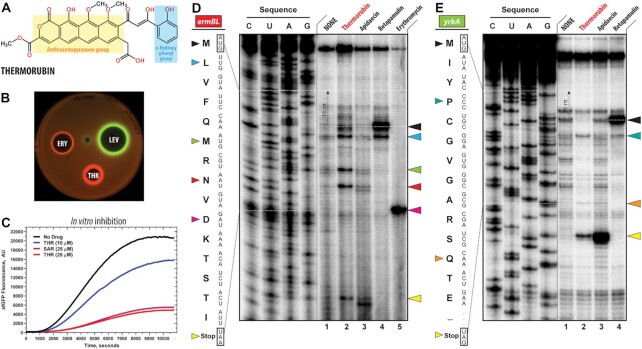
Thermorubin inhibits protein synthesis both in vivo and in vitro. (A) Chemical structure of thermorubin (THR). (B) Induction of a two-color dual reporter system sensitive to inhibitors of the ribosome progression or inhibitors of DNA replication. Spots of erythromycin (ERY), levofloxacin (LEV) and thermorubin (THR) were placed on the surface of an agar plate containing E. coli ΔtolC cells transformed with the pDualrep2 plasmid (14). Induction of expression of Katushka2S (red) is triggered by translation inhibitors, while RFP (green) is induced upon DNA damage. (C) Time-courses of inhibition of sfGFP synthesis by THR (blue and red curves) and sarecycline (SAR, magenta curve) in the in vitro cell-free translation system. AU, arbitrary units. (D, E) Ribosome stalling by THR and other protein synthesis inhibitors on ermBL(D) and yrbA(E) mRNA templates as revealed by reverse transcription inhibition (toe-printing) in a recombinant cell-free translation system. Nucleotide sequences of wild-type ermBL and yrbA genes, and the corresponding amino acid sequence, are shown on the left in each panel. Sequencing lanes (C, U, A, G) are on the left of each gel. Due to the large size of the ribosome, the reverse transcriptase used in the toe-printing assay stops 16 nucleotides downstream of the codon located in the P site. Black arrowhead marks the translation start site (retapamulin control, lane 4). Blue, green, red, teal, and orange arrowheads point to the THR-induced arrest sites within the coding sequences of ermBL or yrbA mRNAs. Magenta arrowhead points to the erythromycin-specific stalling site on ermBL only. Yellow arrowheads point to the translation stop site (apidaecin control, lane 3). Note that, similar to apidaecin, THR induces ribosome stalling at the stop codons of ermBL and yrbA mRNAs. Experiments were repeated twice independently with similar results.