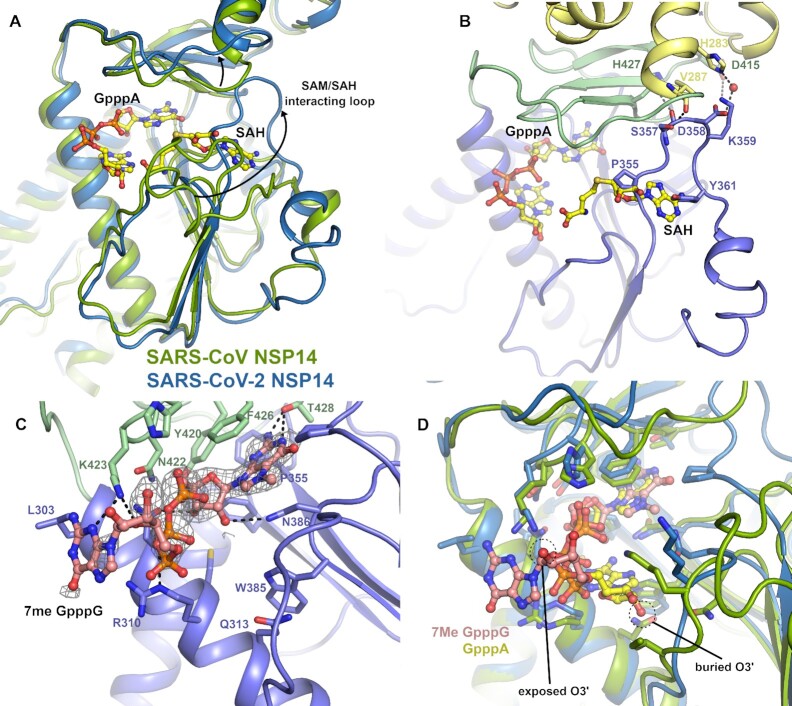
Figure 4.
Structural differences in the NSP14 MTase active site. (A) Alignment of the SARS-CoV NSP14 GpppA/SAH ternary complex (shown in green) with the SARS-CoV-2 NSP14 MTase domain (shown in slate), a significant conformational change can be seen for an extended loop that makes interactions with SAH in the SARS-CoV NSP14 structure. Both ligands are shown in stick format for reference. (B) Close up view of the SAM/SAH interacting loop in SARS-CoV-2 NSP14, residues which overlap with SAH or form contacts to residues in the ExoN or hinge domains are shown in the stick format. (C) Structure of SARS-CoV-2 NSP14 in complex with 7MeGpppG. The 2Fo– 1Fc electron density map is shown in grey in the vicinity of the 7MeGpppG contoured at 1σ. (D) Comparison of the binding poses of the GppA from SARS-CoV NSP14 with the 7MeGpppG from SARS-CoV-2 NSP14. The proteins are colored as for panel A and the positions of the O2′ which is the attachment point for full RNA cap substrates is marked.