FIGURE 6.
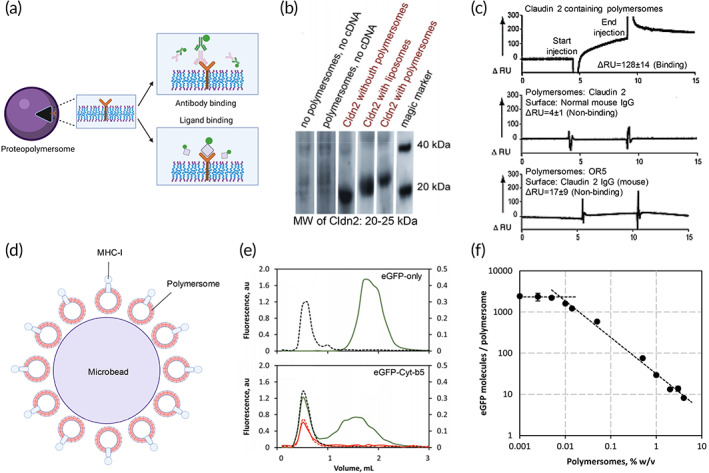
Characterization of membrane receptor‐based proteopolymersomes and their applications. (a) The structure–function relationship of membrane receptors is characterized by binding of conformational antibodies and native ligands to illustrate the proper folding and functions of the receptors, respectively. Schematic was created with BioRender.com. Source: Reproduced with permission from reference 53, Copyright 2012, John Wiley and Sons.(b) Western blot of in vitro expressed claudin‐2 (Cldn2) in the absence or presence of polymersomes or liposomes. (c) SPR measurements showing the binding of Cldn2 reconstituted proteopolymersomes to anti‐Cldn2 functionalized surface but not normal mouse IgG surface. There is also no significant binding between empty polymersomes without Cldn‐2 expression and the anti‐Cldn2 surface.Source: Figure 5b,c is reproduced with permission from reference 52, Copyright 2011, Springer Nature.(d) Engineering of an artificial antigen‐presenting cell by cell‐free in vitro synthesis and incorporation of MHC‐I into polymersomes (red vesicles) and the attachment of MHC‐I proteopolymersomes onto the 3D surface of microbead as a support (purple), forming MHC‐I proteopolymersome‐beads. 111 (e) Size‐exclusion chromatography (SEC) characterization of eGFP (top) and eGFP‐fused transmembrane domain of the rabbit cytochrome b5 (Cyt‐b5) (bottom) proteopolymersomes. Black dashes represent quantification of polymersomes by measuring light absorbance at 350 nm. Green and red solid lines show the presence of eGFP characterized by fluorescence signal (green, made fresh; red, after 6 weeks of storage). (f) There is an inversely proportional dependency of the immobilized eGFP‐Cyt‐b5 molecules per polymersome with increasing polymersome concentration. The polymersome surface area becomes limiting below 0.05% w/v. Source: Figure 5e,f is reproduced with permission from reference 113, Copyright 2016, Springer Nature. Schematics were created with BioRender.com.
