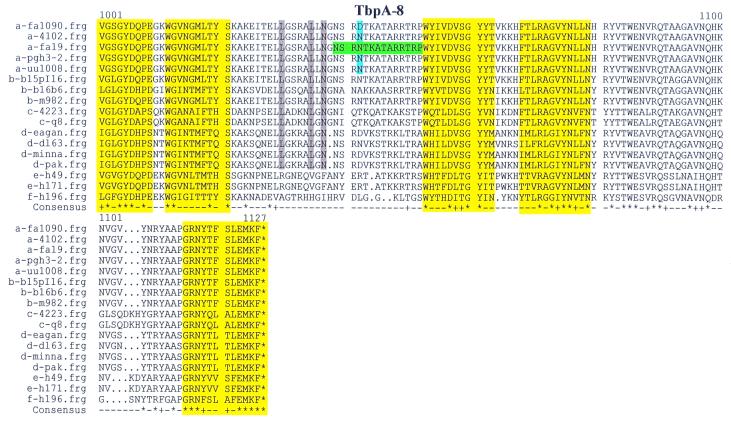
FIG. 1.
Multiple sequence alignment of TbpA proteins. The letter preceding the sequence name to the left of each line indicates the species from which the TbpA sequence was derived: a, N. gonorrhoeae; b, N. meningitidis; c, M. catarrhalis; d, H. influenzae; e, A. pleuropneumoniae; f, P. haemolytica. Boxed single-letter amino acid designations indicate the mature amino terminus of TbpA, if known. Sequences highlighted in green represent the gonococcal strain FA19 TbpA sequences that were synthesized as peptides to generate antipeptide sera. These peptides are numbered and labeled TbpA-1 through TbpA-8, consecutively, from amino terminus to carboxy terminus. Yellow highlighting represents regions homologous to known transmembrane β-strands in E. coli FepA. Blue shading signifies diversity among TbpAs from five gonococcal strains. Gray shading indicates residues that are unique to TbpAs expressed by the human pathogens. Cysteine residues are highlighted in orange. The black vertical arrow represents the carboxy-terminal end of the so-called plug region, defined by homology with E. coli FepA. Dots indicate positions in which gaps were introduced in the alignment; squiggles indicate positions for which no sequence was available. In the consensus line, the asterisks represent complete identity among all 17 aligned sequences; plus signs indicate conservative replacements at that position in the 17-sequence alignment.