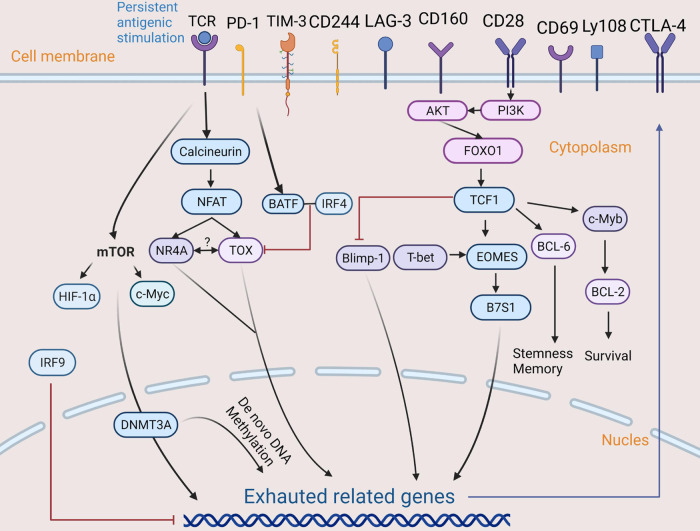
Fig. 3. The complex regulatory network of Tex.
The chronic TCR signal is the core driver of exhaustion, which activates NFAT and its downstream molecules (for example, TOX, NR4A and IRF4) through calcineurin, thereby up-regulating the expression of inhibitory receptors and maintaining the survival of T cells. In the absence of transcription factor AP-1, NFAT activates negative regulatory programs in CD8+ T cells and induces low reactivity in T cells. Then, activation of the secondary transcription factors TOX and NR4A will initiate the CD8+ Tex transcription process. TOX promotes and maintains Tex, while NR4A induces PD-1 and TIM-3 expression and weakens the anti-tumor effect of T cells. BATF is a downstream molecule of PD-1, which can inbibit T cells proliferation, cytokine secretion, and TOX when it combines with IRF4. In addition, transcription factor TCF1 is helpful for maintaining the stem cell-like characteristics of TILs and inducing the transformation of progenitor exhausted T cells. The PI3K/AKT/FOXO1 signaling network is activated by costimulatory molecule CD28. FOXO1 promotes the expression of TCF1, which mediates the transformation of T-bet-to-Eomes transcription factors by promoting Eomes expression in progenitor exhausted CD8+ T cells. TCF1 can also promote the expression of BCL-6 and c-Myb. The latter controls the survival and expression of BCL-2. The overexpression of Eomes induces Tex by activating B7S1. It is worth noting that Blimp-1, like T-bet, can also promote Tex, but Blimp-1 is inhibited by TCF-1. In addition, IRF9 may inhibit the exhaustion of TILs. As for metabolism, mTOR regulates the metabolic checkpoint of glycolysis through transcription factors HIF-1α and c-Myc and participates in methylation-specific exhausted procedures through epigenetic enzymes such as DNMT3A. Moreover, the de novo DNA methyltransferase DNMT3A induces epigenetic silencing of effector and memory-related genes by de novo DNA methylation.