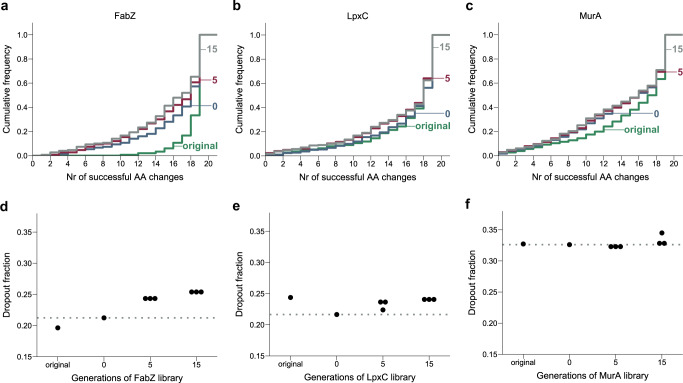
Fig. 2. The stability of library composition across growth cycles varies for different proteins.
The cumulative frequency distribution of successful amino acid substitutions per residue, i.e. edits that could be detected by sequencing, is shown for the FabZ (a), LpxC (b) and MurA (c) libraries. Different curves represent the library composition at different points of sequencing. In case multiple replicates were sequenced (for samples 5 and 15), the first replicate is shown. This figure shows the dropout fraction, i.e. the fraction of edits detected by fewer than 10 reads, in function of the number of generations the FabZ (d), LpxC (e) or MurA (f) libraries were grown. As a reference, the dropout fraction at the start of the selection experiment is indicated by a dotted line. Original refers to the libraries immediately after construction. Generations 0, 5 and 15 refer to the number of generations libraries were grown as part of the selection experiment. As expected, cycles of growth will lead to an increased loss of amino acid substitutions, resulting in flatter cumulative distribution curves and larger dropout fractions. Source data are provided as Supplementary Data 3. AA amino acid.