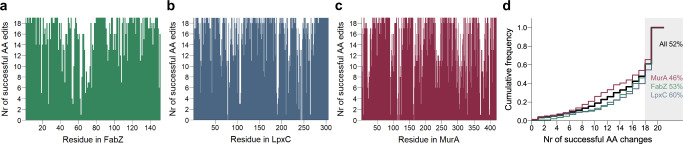
Fig. 3. Saturation editing libraries reveal a surprisingly high tolerance for amino acid substitutions in three essential E. coli proteins.
Bar plots show the number of successful amino acid changes at each position of the proteins FabZ (a), LpxC (b) and MurA (c). Successful amino acid changes were defined as edits that were found to be present in the library after 15 generations of growth (average read count >0). d The cumulative frequency distribution of successful amino acid substitutions detected after 15 generations of growth is shown for all libraries taken together (All) or the FabZ, LpxC and MurA library separately. The percentage of residues that tolerates all or all but one amino acid changes (≥18) is stated and highlighted at the right side of the figure. Source data are provided as Supplementary Data 3. AA amino acid.