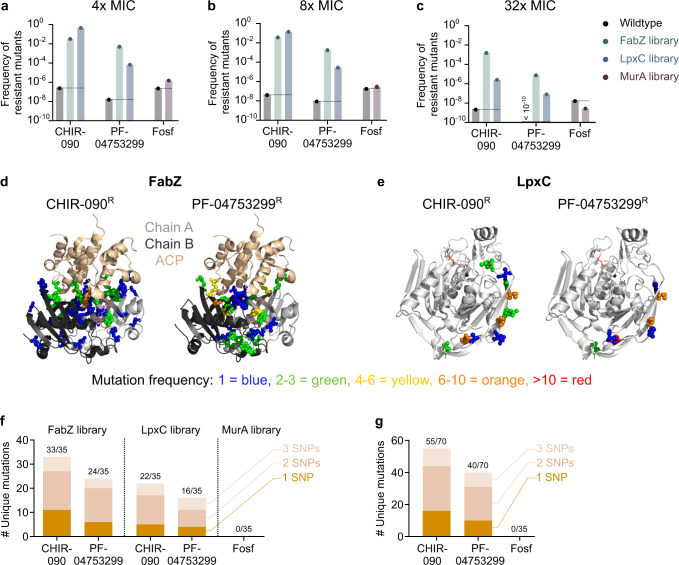
Fig. 7. Saturation editing libraries can guide efforts for the development of novel antibiotics.
a–c The frequency of occurrence of spontaneous resistance mutations is compared to the frequency of occurrence of resistant variants in the saturation editing libraries. This was done by plating either a wild-type culture or the different libraries onto medium containing different concentrations of the compound, i.e. 4x MIC (a), 8× MIC (b) or 32× MIC (c), and counting the number of colonies that developed after overnight growth. These numbers were normalized to the total cell numbers present in the wild-type culture or the libraries, respectively. d The location of targeted residues in the FabZ protein is shown for both CHIR-090 and PF-04753299. Residues are colored according to the number of times they were targeted in isolated resistant variants. Only one dimer of the FabZ hexamer is shown for clarity. Mutations are indicated in both chain A and B. e The location of targeted residues in the LpxC protein is shown for both CHIR-090 and PF-04753299. Residues are colored according to the number of times they were targeted in isolated resistant variants. The number of unique mutations in fabZ and lpxC that provide resistance to CHIR-090 or PF-04753299 and the number of mutations in murA that provide resistance to fosfomycin are shown, either grouped per library and compound (f) or grouped per compound only (g). For each library-compound combination, 35 resistant variants were isolated and their fabZ, lpxC or murA gene was sequenced. The mutations found are subdivided into categories based on the minimal number of SNPs necessary to provide the observed amino acid change. Source data are provided as a Source Data file. Fosf fosfomycin, ACP acyl carrier protein.