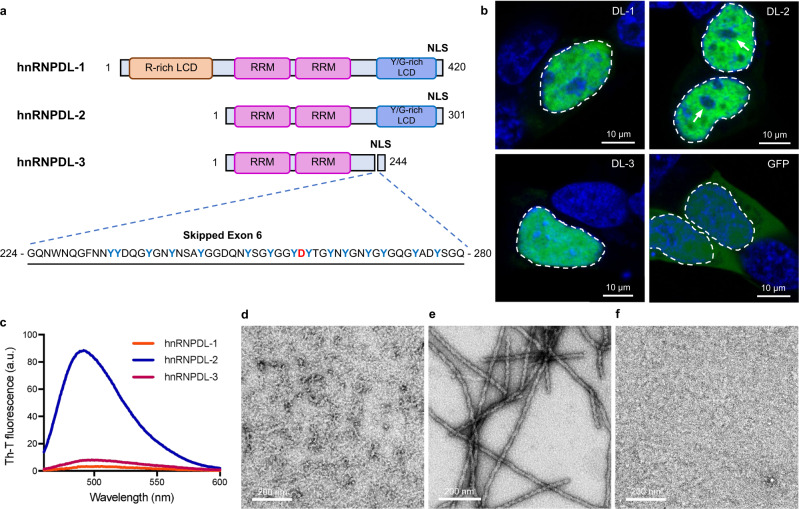
Fig. 1. hnRNPDL-2 aggregates into ordered amyloid fibrils.
a Domain organization of human hnRNPDL isoforms. The RNA recognition motifs are labelled as RRM (pink) and N- and C-terminal low complexity domains (LCD) are indicated as R-rich LCD (orange) and Y/G-rich LCD (blue), respectively. The region and sequence of exon 6 is also indicated, according to hnRNPDL-2 numbering. D259 disease-associated amino acid is shown in red. NLS indicates the shared nuclear localization signal. b Representative confocal images of HeLa hnRNPDL KO cells transiently transfected with the different hnRNPDL isoforms (DL-1, DL-2 and DL-3). The empty GFP vector (GFP) was transfected as control. The images depict the characteristic nuclear distribution of GFP-hnRNPDL fusions (green). Nuclear DNA was stained with Hoechst (blue). White arrows indicate the location of the nucleolus. In all cases, the nucleus contour has been indicated with a white dashed line. Scale bar, 10 μm. c Th-T binding to hnRNPDL-1 (orange line), hnRNPDL-2 (blue line) and hnRNPDL-3 (purple line) after incubation at 37 °C and 600 rpm, pH 7.5 and 300 mM NaCl for 2 days. Representative negative staining TEM micrographs of the incubated solutions of (d) hnRNPDL-1, (e) hnRNPDL-2 and (f) hnRNPDL-3. Scale bar, 200 nm. Note that only in (e) individual amyloid filaments are evident. In (b) and (d–f), results are representative from three independent experiments.