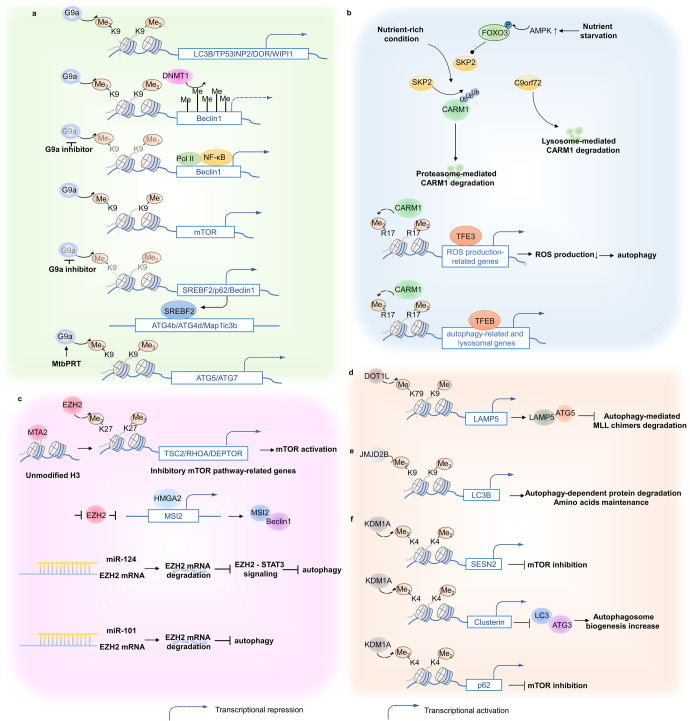
Fig. 2.
Histone methylation and autophagy regulation. a G9a negatively regulates core autophagy effectors and upstream autophagic regulators to influence autophagy level indirectly. b Under a nutrient-rich environment, SKP2 mediates CARM1 protein degradation, nutrient starvation activates AMPK-dependent FOXO3 phosphorylation, which transcriptionally represses SKP2, resulting in CARM1-mediated transcriptional activation of autophagy-related and lysosomal genes. c EZH2 repressively regulates autophagy via mTOR activation. EZH2 negatively controls TSC2/RHOA/DEPTOR gene transcription to elicit mTOR pathway, leading to autophagy inhibition. Additionally, EZH2 represses HMGA2 expression. HMGA2 can directly activate the MSI2 promoter region, which triggers autophagy via Beclin1 interactions. d DOT1L elevates LAMP5 expression via H3K79 methylation modification enhancements. LAMP5 directly interacts with ATG5 to interrupt an autophagy flux, protecting MLL chimeras from autophagy degradation. e JMJD2B promotes autophagy occurrence via histone demethylation at LC3B promoters, assisting in intracellular amino acid maintenance. f KDM1A negatively manipulates autophagy flux through SESN2- and CLU-dependent pathways or directly targeting p62. SESN2 inhibits mTORC1 activity, and CLU increases autophagosome biogenesis through MAP1LC3/LC3-ATG3 heterodimer stabilization