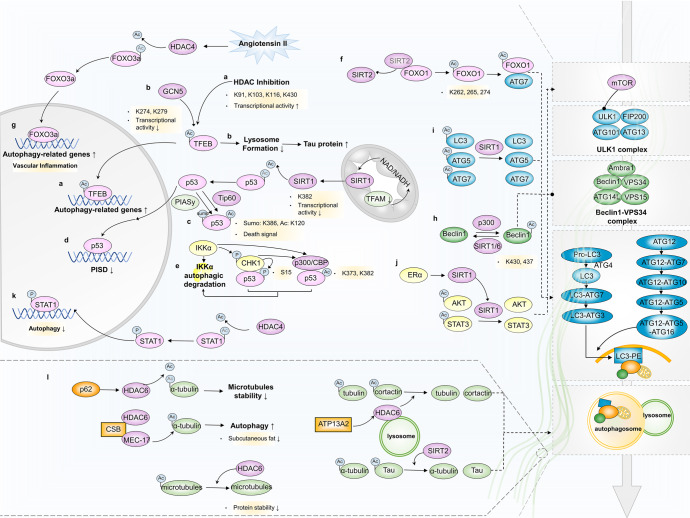
Fig. 5.
Acetylation modification and autophagy regulation. a TFEB acetylation at K91, K103, K116, and K430 markedly activates the expression of its target genes related to autophagy and lysosomal biogenesis and induces cell death. b GCN5-catalyzed K274 and K279 acetylation of TFEB impedes lysosome formation and the clearance of Tau protein aggreagates. c Tip60-mediated p53 acetylation at K120 and K386 sumoylation function as a death signal to promote p53 cellular accumulation and autophagy. d P53 acetylation at K382 sites affects autophagy levels via the transcriptional regulation of the PISD enhancer. e P53 is phosphorylated by CHK1 activation and undergoes p300/CEP-mediated acetylation at the K373 and/or K382 sites, which triggers autophagy and autophagy-mediated IKKα degradation. f Cytoplasmic FOXO1 is acetylated at K262, K265, and K274 via SIRT2 dissociation, facilitating the interaction between cytosolic FOXO1 and ATG7 and the autophagic process. g Upregulated HDAC4 deacetylates FOXO3a to transcriptionally activate autophagy. h P300 and SIRT1/6-regulated acetylation of Beclin1 at the K430 and K437 sites. i SIRT1 overexpression deacetylates LC3, ATG5 and ATG7 and activates autophagy. j ERα-induced SIRT1 expression deacetylates and activates AKT and STAT3, resulting in suppression of autophagy via mTOR-ULK1 and p55 cascade. k HDAC4-STAT1 signaling-regulating autophagy inhibition. (l) HDAC6-mediated cytoskeleton proteins acetylation regulation