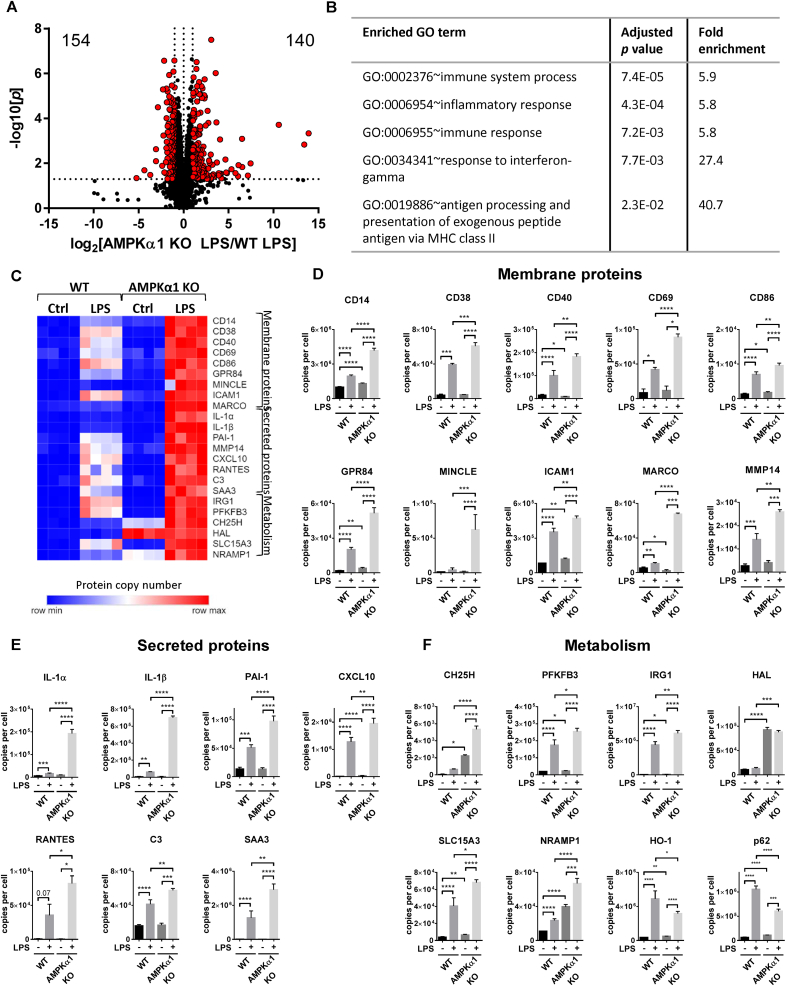
Figure 1.
Deletion of AMPKα1 drives dynamic changes in the proteome of LPS-stimulated macrophages. (A) Volcano plot shows log2 fold change in mean protein copy numbers per cell between WT and AMPKα1 KO BMDMs stimulated for 24 h with 100 ng/ml LPS. Proteins highlighted in red have a significant (p < 0.05) fold change <0.5 or >2. (B) The top five enriched biological process GO terms for proteins expressed >2-fold higher (p < 0.05) in LPS-stimulated macrophages lacking AMPKα1. (C) Heat maps showing proteins associated with M1 macrophages in control and LPS-stimulated BMDMs from WT or AMPKα1 KO mice. Relative protein abundance is graded from low (blue) to high (red). (D–F) Mean protein copy number per cell are shown for (D) membrane proteins (CD14, CD38, CD40, CD69, CD86, GPR84, MINCLE, ICAM-1, MARCO and MMP-14), (E) secreted proteins (IL-1α, IL-1β, PAI-1, CXCL10, RANTES, C3 and SAA3) and (F) proteins implicated in metabolism (CH25H, PFKFB3, IRG1, HAL, SLC15A3, NRAMP1, HO-1 and p62). Bar charts represent the mean and standard deviation of results from biological quadruplicates. P values were calculated as described in Methods; a p < 0.05 is represented by ∗, p < 0.01 by ∗∗, p < 0.001 by ∗∗∗, and p < 0.0001 by ∗∗∗∗. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)