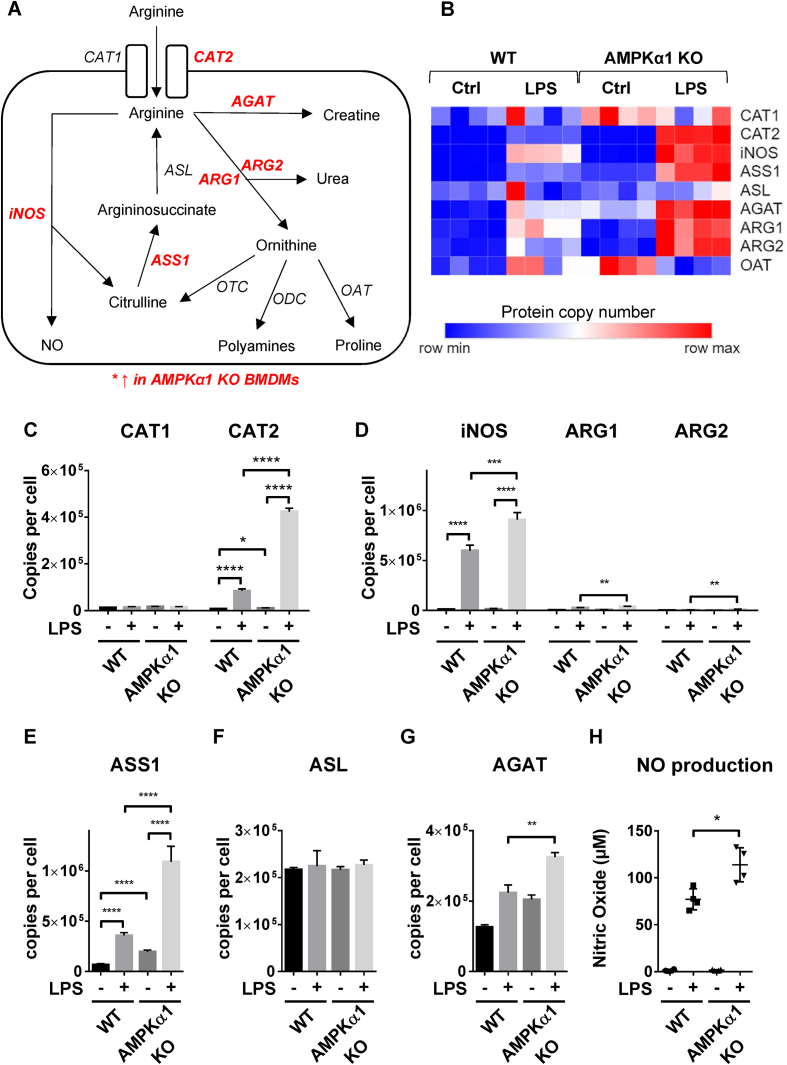
Figure 3.
Arginine metabolic enzymes are enhanced in AMPKα1 KO macrophages. (A) Schematic of arginine metabolism in macrophages. Proteins with enhanced expression in AMPKα1 KO BMDMs are shown in red italics. (B) Heat maps of proteins associated with arginine metabolism in control and LPS-stimulated BMDMs from WT or AMPKα1 KO mice. Relative protein abundance is graded from low (blue) to high (red). (C–G) Mean protein copy number per cell are shown for (C) CAT1 and CAT2, (d) iNOS, ARG1 and ARG2, (e) ASS1, (f) ASL, and (G) AGAT. Bar charts represent the mean and standard deviation of results from biological quadruplicates. P values were calculated as described in Methods; a p < 0.05 is represented by ∗, p < 0.01 by ∗∗, p < 0.001 by ∗∗∗, and p < 0.0001 by ∗∗∗∗. (H) WT or AMPKα1 KO BMDMs were stimulated ±100 ng/ml LPS for 24 h, and the levels of nitric oxide (NO) secreted into the media were determined as described in Methods. Graphs represent the mean and standard deviation of results from 4 biological replicates. A p < 0.05 is indicated by ∗ (two-tailed Student’s t-test assuming unequal variance). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)