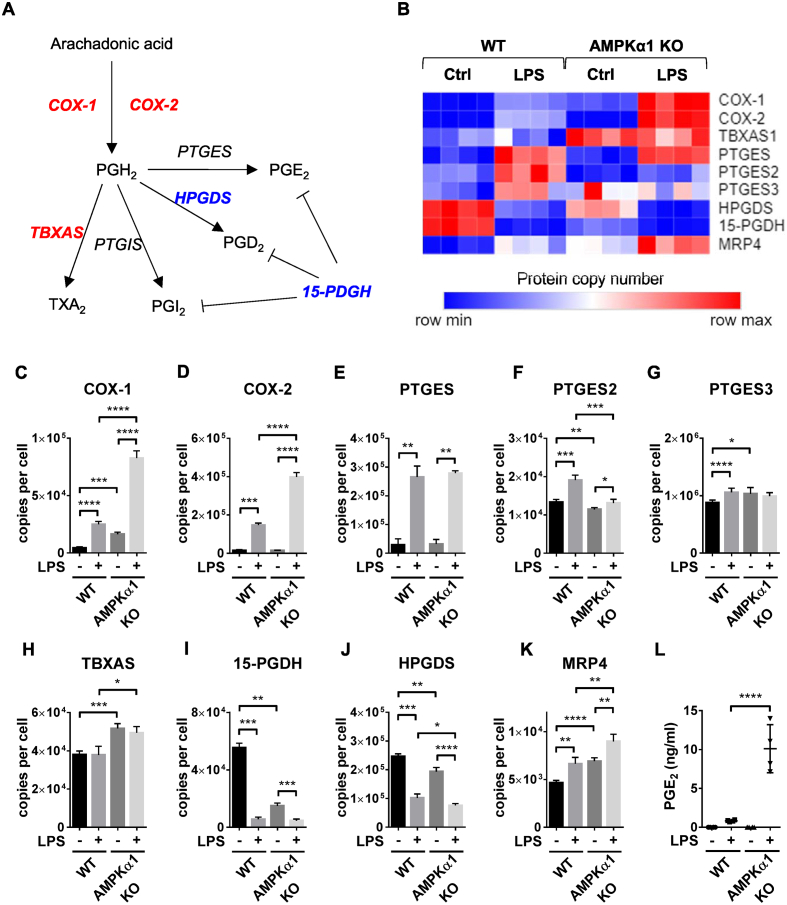
Figure 4.
AMPK regulates prostaglandin synthesis in inflammatory macrophages. (A) Schematic of prostaglandin synthesis in macrophages. COX-1/2 are rate-limiting enzymes that catalyse the conversion of arachidonic acid to PGH2. PGH2 is subsequently converted to PGE2 by prostaglandin E synthase (PTGES), PGD2 by prostaglandin D synthase (PGDS), PGI2 by prostaglandin I synthase (PTGIS), and TXA2 by thromboxane synthase (TBXAS). Prostaglandin dehydrogenase (15-PGDH) plays a key role in the degradation of prostaglandins. Proteins with increased expression in AMPKα1 KO BMDMs are shown in red, and those with decreased expression are shown in blue. (B) Heat maps of proteins associated with prostaglandin synthesis in control and LPS-stimulated BMDMs from WT or AMPKα1 KO mice. Relative protein abundance is graded from low (blue) to high (red). (C–K) Mean protein copy number per cell are shown for (C) COX-1, (D) COX-2, (E) TBXAS, (F) PTGES, (G) PTGES2, (H) PTGES3, (I) HPGDS, (J) 15-PGDH and (K) MRP4. Bar charts represent the mean and standard deviation of results from biological quadruplicates. P values were calculated as described in Methods; a p < 0.05 is represented by ∗, p < 0.01 by ∗∗, p < 0.001 by ∗∗∗, and p < 0.0001 by ∗∗∗∗. (L) BMDMs from WT or AMPKα1 KO mice were stimulated with 100 ng/ml LPS for 24 h, and the levels of PGE2 secreted into the media were determined as described in Methods. Graphs represent the mean and standard deviation of results from 4 biological replicates. Two-way ANOVA indicated a significant effect of genotype on PGE2 levels (F = 36.32, p < 0.0001). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)