FIGURE 1.
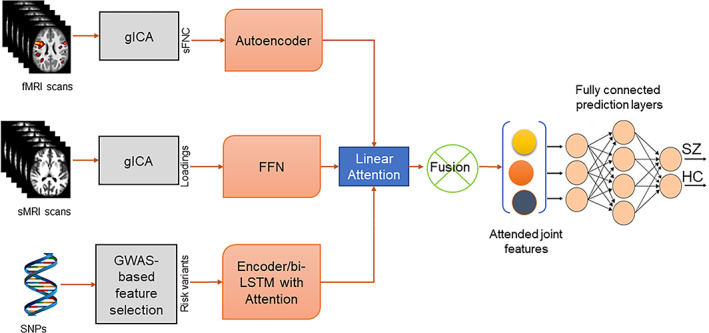
Our multimodal architecture. It has two submodules, (i) group independent component analysis (gICA) and genetic variable selection (ii) deep neural networks (DNN) for learning the modality features. The submodule (i) runs two separate gICA for structural and functional magnetic resonance imaging (sMRI and (fMRI) with distinct settings, for example, number of independent components expected. Then it computes the static functional network connectivity (sFNC) matrix for each subject from fMRI gICA and collects the ICA loading matrix from sMRI. Then, the genotyping on the genomics sequence generates single nucleotide polymorphism (SNPs). The DNN submodule consists of four subnetworks. sFNC features are extracted and learned by an AE with mean square error loss. The next subnetwork is a multilayer feed forward network for learning the ICA loading features. For genomics, we use a bi‐directional long short‐term memory (LSTM) unit with attention mechanism. After that, a fusion mechanism is applied to combine the latent features from three modalities using an attention model. The latent features are weighted by the attention score. Finally, the joint features are sent through a series of fully connected layers followed by a SoftMax prediction layer. The overall model is optimized using an Adam optimizer.
