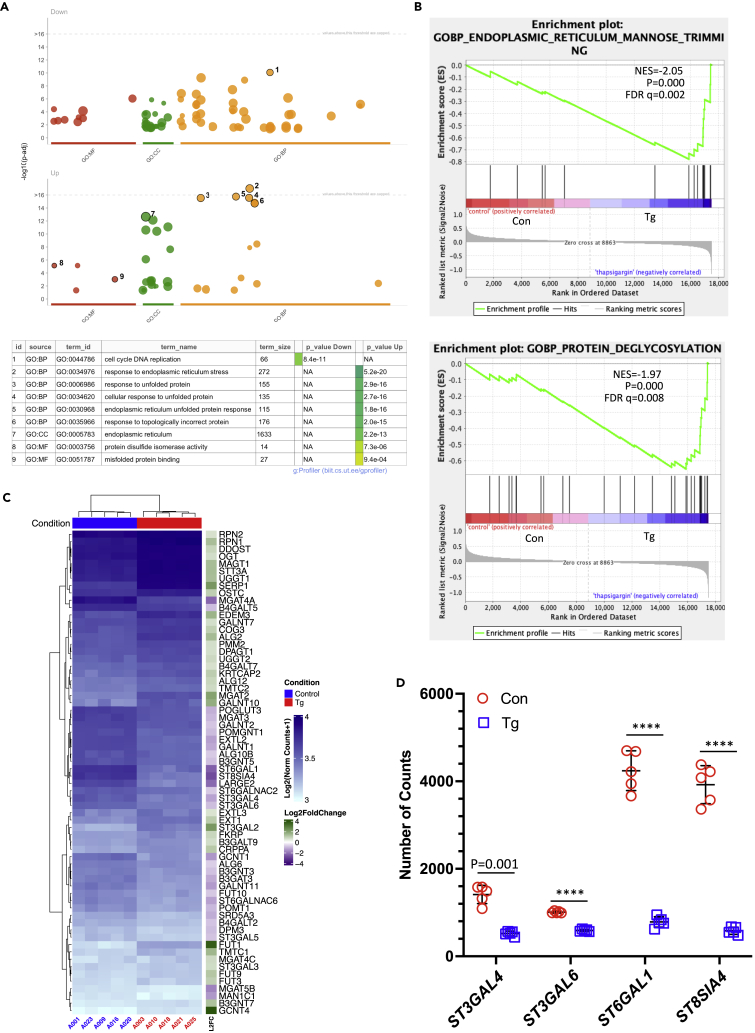
Figure 2.
Genomic analysis in BeWo cells after Tg treatment
RNA-Seq was used to analyze transcript changes induced by Tg in BeWo cells.
(A) g.Profiler analysis with GO_BP revealed biological processes affected by thapsigargin treatment.
(B) GSEA with GO_BP showed enrichment of transcripts required for Mannose Trimming, a key process in glycosylation and Protein Deglycosylation.
(C) Heatmap of 66 differentially expressed genes (FC > 1.5, Padj < 0.05) induced by Tg treatment involved in protein glycosylation from gene ontology (GO: 0006486).
(D) Sialyltransferases involved in N-glycan sialylation are down-regulated in Tg-treated BeWo cells. The number of normalized counts from RNA-Seq is plotted. Data are mean ± SD, n = 5. Paired t-test. ∗∗∗∗ indicates p < 0.0001. NES, normalized enrichment score; Con, control; Tg, Thapsigargin.